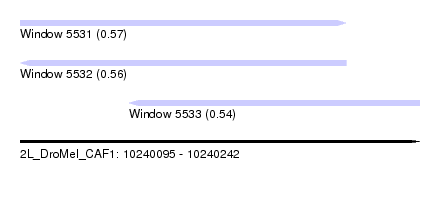
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,240,095 – 10,240,242 |
| Length | 147 |
| Max. P | 0.568086 |

| Location | 10,240,095 – 10,240,215 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -29.90 |
| Consensus MFE | -25.22 |
| Energy contribution | -25.02 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568086 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10240095 120 + 22407834 UUGCAUUUCAGCGCCAUGAACAGUUAUUCGUGGGCUGCCAUUUUAAAAUUUGGAGUAGCGUUUCUUUUUCUCUCCUCCAUAAAUGUGCGAAAGAGAAAAAAUGUUGAGUUCAAUCGCUUU ..((....((((.(((((((......)))))))))))............(((((.((((((((...(((((((..(((((....))).)).)))))))))))))))..)))))..))... ( -32.00) >DroSec_CAF1 4423 120 + 1 UUCCAUUUCAGCGCCGUGAACAGUUAUUCGUGGGCUGCCAUUUUAAAAUUUGGAGUAGCGUUUCUUUUUCUCUCCUUCAUAAAAGUGCGAAAGAGAAAAAAUGUUGAGUUCAAUCGCAUU ..........(((...(((((.(((((((((((....))))...........))))))).((((((((((.(..(((.....))).).)))))))))).........)))))..)))... ( -27.30) >DroSim_CAF1 4857 120 + 1 UUGCAUUUCAGCGCCGUGAACAGUUAUUCGUGGGCUGCCAUUUUAAAAUUUGGAGUAGCGUUUCUUUUUCUCUCCUUCAUAAAAGUGCGAAAGAGAAAAAAUGUUGAGUUCAAUCGCAUU .(((....((((.(((((((......)))))))))))............(((((.((((((((...(((((((....(((....)))....)))))))))))))))..)))))..))).. ( -28.10) >DroEre_CAF1 4356 120 + 1 UUGCACUUCAGCAGCGUGAACAGUUAUUCGUGGGCUGCCAUUUUAAAAUUUGGAGUAGCGUUUCUUUUUCUCUGCUCCAUAAAAGUGCGAAAGAGAAAAGAUGUUACGUCCAACCGCUUU ..((......(((((.((((......))))...)))))...........(((((((((((((...((((((((..(((((....))).)).)))))))))))))))).)))))..))... ( -33.60) >DroYak_CAF1 4232 120 + 1 UUGCAUUUCAGCAGCGUGAACAGUUAUUCGUGGGCUGCCAUUUUAAAAUUUGGAGUAGCGUUUCUUUUUCUCUGCUCCACUAAAGUGCGAAAGAGAAAAGAUCUUAAGUUUAGUAGCUUU .........(((.((.(((((.(((((((((((....))))...........))))))).....(((((((((..(((((....))).)).))))))))).......))))))).))).. ( -28.50) >consensus UUGCAUUUCAGCGCCGUGAACAGUUAUUCGUGGGCUGCCAUUUUAAAAUUUGGAGUAGCGUUUCUUUUUCUCUCCUCCAUAAAAGUGCGAAAGAGAAAAAAUGUUGAGUUCAAUCGCUUU ..........(((...(((((.(((((((((((....))))...........))))))).....(((((((((..(((((....))).)).))))))))).......)))))..)))... (-25.22 = -25.02 + -0.20)

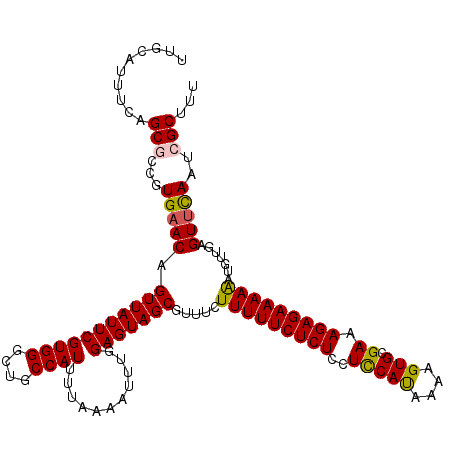
| Location | 10,240,095 – 10,240,215 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -20.40 |
| Energy contribution | -21.08 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.556796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10240095 120 - 22407834 AAAGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACAUUUAUGGAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAAUGGCAGCCCACGAAUAACUGUUCAUGGCGCUGAAAUGCAA ..((((.(((....)))..((((((((((((((........)))))))))))))).....))))...............((..(((((((.((((....)))).))).))))..)).... ( -31.60) >DroSec_CAF1 4423 120 - 1 AAUGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACUUUUAUGAAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAAUGGCAGCCCACGAAUAACUGUUCACGGCGCUGAAAUGGAA ...(((.(((....)))..((((((((((((((........)))))))))))))).....)))...((((.(((....)))..((((((..((((....))))..)).))))...)))). ( -28.50) >DroSim_CAF1 4857 120 - 1 AAUGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACUUUUAUGAAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAAUGGCAGCCCACGAAUAACUGUUCACGGCGCUGAAAUGCAA ...(((.(((....)))..((((((((((((((........)))))))))))))).....)))................((..((((((..((((....))))..)).))))..)).... ( -27.70) >DroEre_CAF1 4356 120 - 1 AAAGCGGUUGGACGUAACAUCUUUUCUCUUUCGCACUUUUAUGGAGCAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAAUGGCAGCCCACGAAUAACUGUUCACGCUGCUGAAGUGCAA ((((.((..(((.......)))..)).)))).(((((((..(((((.((.(.........).)).)))))...........((((((....((((....))))..))))))))))))).. ( -28.50) >DroYak_CAF1 4232 120 - 1 AAAGCUACUAAACUUAAGAUCUUUUCUCUUUCGCACUUUAGUGGAGCAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAAUGGCAGCCCACGAAUAACUGUUCACGCUGCUGAAAUGCAA ...((..............(((((((((((((.(((....)))))).)))).)))))).......................((((((....((((....))))..)))))).....)).. ( -25.40) >consensus AAAGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACUUUUAUGGAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAAUGGCAGCCCACGAAUAACUGUUCACGGCGCUGAAAUGCAA ...(((..(((((......((((((((((((((........))))))))))))))......(((...(((..........))).)))............)))))...))).......... (-20.40 = -21.08 + 0.68)

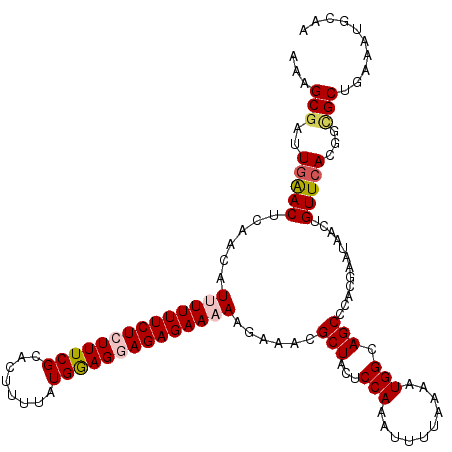
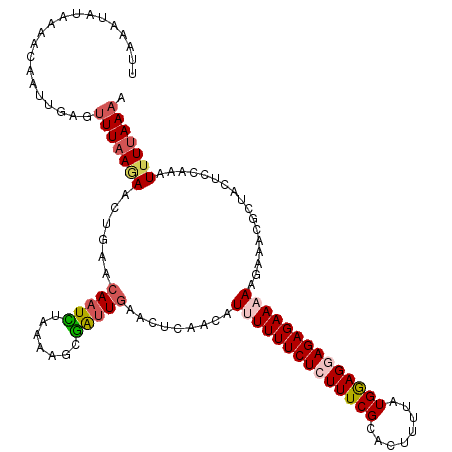
| Location | 10,240,135 – 10,240,242 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.92 |
| Mean single sequence MFE | -21.61 |
| Consensus MFE | -13.12 |
| Energy contribution | -13.96 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.543707 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10240135 107 - 22407834 UUGAAUAU-------------UUAAGAACUGAACAAUCUAAAAGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACAUUUAUGGAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAA .......(-------------((((((..((..(((((.......))))).........((((((((((((((........)))))))))))))).............))..))))))). ( -19.90) >DroSec_CAF1 4463 120 - 1 CUAAGUAUUAAACAAUUGAGUUUAAGAACUGAACAAUCUAAAUGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACUUUUAUGAAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAA ...((((........((((((((((......................))))))))))..((((((((((((((........))))))))))))))........))))............. ( -22.95) >DroSim_CAF1 4897 120 - 1 CUAAAUAUUAAACAAUUGAGUUUUAGAACUGAACAAUCUAAAUGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACUUUUAUGAAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAA .......(((((.....((((((((((.........)))))).(((.(((....)))..((((((((((((((........)))))))))))))).....))).)))).....))))).. ( -22.90) >DroEre_CAF1 4396 120 - 1 UUAAAUAUAUAAAAAUACCGUUUAAAAACGGAGCAAUUUAAAAGCGGUUGGACGUAACAUCUUUUCUCUUUCGCACUUUUAUGGAGCAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAA .................(((((....))))).....(((((((....(((((.(((.(.((((((.(((((.((.((.....)).)))))))))))))...).)))))))).))))))). ( -24.30) >DroYak_CAF1 4272 120 - 1 UUAAAUAUACAAUAAUCGAGUUUAAUAACUGGGCAAUAUAAAAGCUACUAAACUUAAGAUCUUUUCUCUUUCGCACUUUAGUGGAGCAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAA .................(((((((.......(((.........)))..)))))))....(((((((((((((.(((....)))))).)))).))))))...................... ( -18.00) >consensus UUAAAUAUAAAACAAUUGAGUUUAAGAACUGAACAAUCUAAAAGCGAUUGAACUCAACAUUUUUUCUCUUUCGCACUUUUAUGGAGGAGAGAAAAAGAAACGCUACUCCAAAUUUUAAAA ....................(((((((......(((((.......))))).........((((((((((((((........)))))))))))))).................))))))). (-13.12 = -13.96 + 0.84)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:54:02 2006