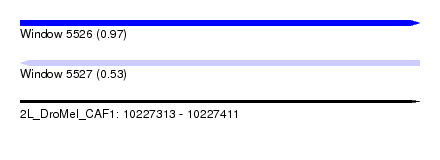
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,227,313 – 10,227,411 |
| Length | 98 |
| Max. P | 0.968751 |

| Location | 10,227,313 – 10,227,411 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -36.27 |
| Consensus MFE | -24.17 |
| Energy contribution | -26.37 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.968751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
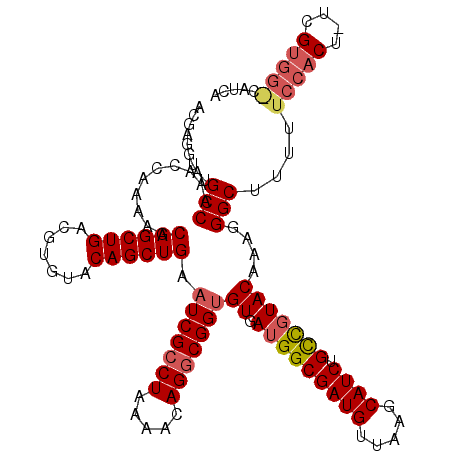
>2L_DroMel_CAF1 10227313 98 + 22407834 ACUAGAUAUGCCAAACCAAAAGCAGCUGACGCGUACAGCUGAAUCGCCUAAAAUAGGCGGGGUGAUGG-----UUAA-------CUCUACAAAGGGCCAAGCCCACG-UUG--------- ..((((...((((.(((.....((((((.......))))))..(((((((...))))))))))..)))-----)...-------.))))....((((...))))...-...--------- ( -33.40) >DroSec_CAF1 2015 120 + 1 ACGAGGUAUGCCAAACCAAAAGCAGCUGACGUGUACAGCUGAAUCGACUGAAACAGGCGGUGUGAUGGCGAUGUUAAGCAUCUGUCGUACAUAGGGCUUUUUCCACUUUCGUGGACAUCA .((((((.......))).....((((((.......))))))..)))...((...((((..(((((((((((((.....)))).))))).))))..))))..(((((....)))))..)). ( -34.40) >DroSim_CAF1 2270 119 + 1 ACGAGGUAUGCCAAACCAAAAGCAGCUGACGUGUACAGCUGAAUCGCCUAAAACAGGCGGUGUGAUGGCGAUGUUAAGCAUCUGCCGUACAAAUGGCUUUUUCCACU-UCGUGGGCAUCA (((((((..((((.........((((((.......)))))).(((((((.....)))))))....))))((((.....)))).(((((....))))).......)))-))))........ ( -41.00) >consensus ACGAGGUAUGCCAAACCAAAAGCAGCUGACGUGUACAGCUGAAUCGCCUAAAACAGGCGGUGUGAUGGCGAUGUUAAGCAUCUGCCGUACAAAGGGCUUUUUCCACU_UCGUGG_CAUCA .........(((..........((((((.......)))))).(((((((.....)))))))((.(((((((((.....)))).)))))))....)))....(((((....)))))..... (-24.17 = -26.37 + 2.20)

| Location | 10,227,313 – 10,227,411 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.88 |
| Mean single sequence MFE | -33.37 |
| Consensus MFE | -22.81 |
| Energy contribution | -24.37 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.68 |
| SVM decision value | -0.00 |
| SVM RNA-class probability | 0.533546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
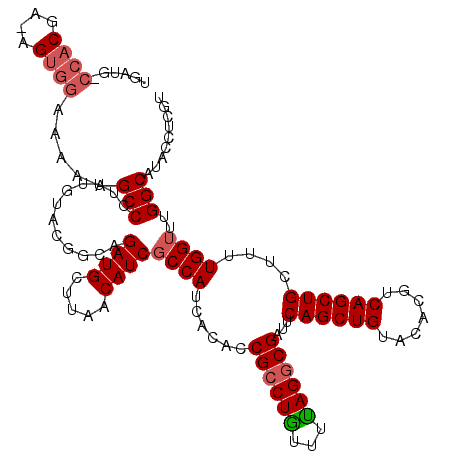
>2L_DroMel_CAF1 10227313 98 - 22407834 ---------CAA-CGUGGGCUUGGCCCUUUGUAGAG-------UUAA-----CCAUCACCCCGCCUAUUUUAGGCGAUUCAGCUGUACGCGUCAGCUGCUUUUGGUUUGGCAUAUCUAGU ---------...-...((((...))))....((((.-------....-----(((..(((.((((((...))))))...((((((.......)))))).....))).)))....)))).. ( -28.80) >DroSec_CAF1 2015 120 - 1 UGAUGUCCACGAAAGUGGAAAAAGCCCUAUGUACGACAGAUGCUUAACAUCGCCAUCACACCGCCUGUUUCAGUCGAUUCAGCUGUACACGUCAGCUGCUUUUGGUUUGGCAUACCUCGU .(((((((((....)))))..((((....((.....))...))))..))))((((...((.((.(((...))).))...((((((.......))))))....))...))))......... ( -34.30) >DroSim_CAF1 2270 119 - 1 UGAUGCCCACGA-AGUGGAAAAAGCCAUUUGUACGGCAGAUGCUUAACAUCGCCAUCACACCGCCUGUUUUAGGCGAUUCAGCUGUACACGUCAGCUGCUUUUGGUUUGGCAUACCUCGU ...((((.((.(-(((((......))))))))..))))((((.....))))((((...((.((((((...))))))...((((((.......))))))....))...))))......... ( -37.00) >consensus UGAUG_CCACGA_AGUGGAAAAAGCCCUUUGUACGGCAGAUGCUUAACAUCGCCAUCACACCGCCUGUUUUAGGCGAUUCAGCUGUACACGUCAGCUGCUUUUGGUUUGGCAUACCUCGU ......((((....)))).....(((............((((.....))))((((......((((((...))))))...((((((.......))))))....))))..)))......... (-22.81 = -24.37 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:56 2006