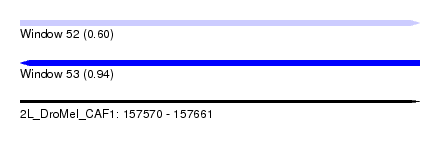
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 157,570 – 157,661 |
| Length | 91 |
| Max. P | 0.943219 |

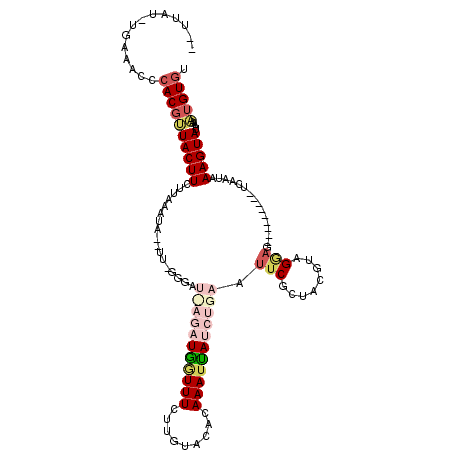
| Location | 157,570 – 157,661 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -4.98 |
| Energy contribution | -7.30 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.601538 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
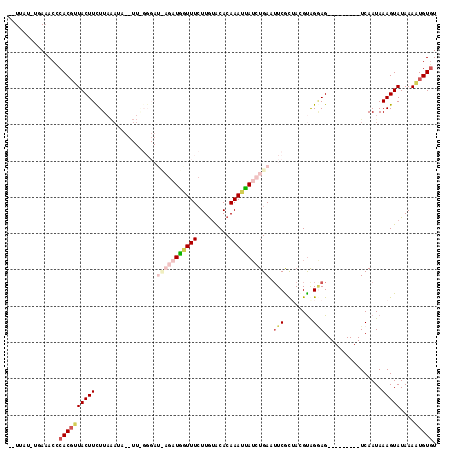
>2L_DroMel_CAF1 157570 91 + 22407834 --UUAU-UGAAACCCACGUUACUUCUUAGAU---------------UAGUUUGUUGUGCACAAAUUAUCUGAAUUCGCUAGGUGGAAG---------UCAAUAAAGUAUAAAAUGUGU --((((-(((...((((.(......((((((---------------.(((((((.....))))))))))))).......).))))...---------))))))).............. ( -20.02) >DroSec_CAF1 4016 106 + 1 --UUAU-UGAAACCCACGUUACUUCUUAAAUAUUUUAGGGAUCAGAUGGUUUCUUGUACACAAAUUAUCUGAAUUCGCGACGUAGGAG---------UCAAUAAAGUAUAAAAUGUGU --((((-(((..((.((((..((((((((.....)))))))(((((((((((.........)))))))))))....)..)))).))..---------))))))).............. ( -26.60) >DroSim_CAF1 3935 106 + 1 --UUAU-UGAAACCCACGUUACUUCUUAAAUAUUUUGGGGAUCAGAUGGUUUCUUGUACACAAAUUAUCUGAAUUCGUGACGUAGGAG---------UCAAUAAAGUAUAAAAUGUGU --((((-(((..((.((((((((((((((.....)))))))(((((((((((.........)))))))))))....))))))).))..---------))))))).............. ( -29.80) >DroEre_CAF1 3992 92 + 1 AGUUAGUUUAAACUCACUAUACUUCUU-----------------GAUGAUUUCUUGAACACAAAAUAAGUGAAUUCACUAUGUGGCAG---------UCAAUAAAGUAUAAAAUGUGU .((.(((....))).))(((((((.((-----------------(((...........((((.....((((....)))).))))...)---------))))..)))))))........ ( -12.54) >DroYak_CAF1 3948 117 + 1 ACUUAAUUUA-ACUCACGAUACUUUUUAAAUAGAUUGGGGAUUAGAUGAUUUCUUAUGCACAAAUCAUCUGAAGUCGCUAGGCAGGAGUCGCUAACGUCAAUAAAGUAUAAAUUGUUU .........(-((.....(((((((.......(.((((.(((((((((((((.........))))))))))).(((....))).....)).)))))......))))))).....))). ( -22.12) >consensus __UUAU_UGAAACCCACGUUACUUCUUAAAUA__UU_GGGAU_AGAUGGUUUCUUGUACACAAAUUAUCUGAAUUCGCUACGUAGGAG_________UCAAUAAAGUAUAAAAUGUGU ..............((((((((((.................(((((((((((.........))))))))))).(((........)))................)))))....))))). ( -4.98 = -7.30 + 2.32)

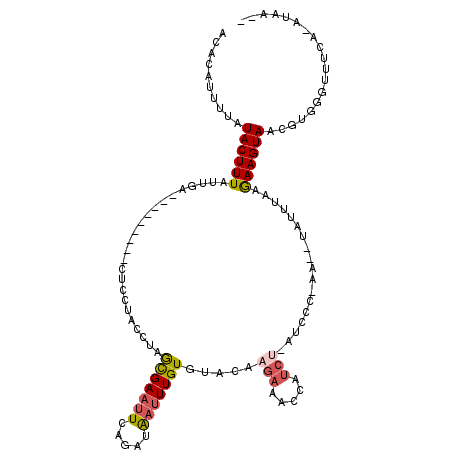
| Location | 157,570 – 157,661 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.83 |
| Mean single sequence MFE | -19.92 |
| Consensus MFE | -2.62 |
| Energy contribution | -3.18 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.13 |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.943219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 157570 91 - 22407834 ACACAUUUUAUACUUUAUUGA---------CUUCCACCUAGCGAAUUCAGAUAAUUUGUGCACAACAAACUA---------------AUCUAAGAAGUAACGUGGGUUUCA-AUAA-- ..............(((((((---------..(((((...((...((.((((..(((((.....)))))...---------------))))))...))...)))))..)))-))))-- ( -17.60) >DroSec_CAF1 4016 106 - 1 ACACAUUUUAUACUUUAUUGA---------CUCCUACGUCGCGAAUUCAGAUAAUUUGUGUACAAGAAACCAUCUGAUCCCUAAAAUAUUUAAGAAGUAACGUGGGUUUCA-AUAA-- ..............(((((((---------..(((((((..(....((((((..(((.(.....).)))..))))))...((..........))..)..)))))))..)))-))))-- ( -23.00) >DroSim_CAF1 3935 106 - 1 ACACAUUUUAUACUUUAUUGA---------CUCCUACGUCACGAAUUCAGAUAAUUUGUGUACAAGAAACCAUCUGAUCCCCAAAAUAUUUAAGAAGUAACGUGGGUUUCA-AUAA-- ..............(((((((---------..(((((((.((...((.(((((.((((......(((.....)))......)))).)))))))...)).)))))))..)))-))))-- ( -22.60) >DroEre_CAF1 3992 92 - 1 ACACAUUUUAUACUUUAUUGA---------CUGCCACAUAGUGAAUUCACUUAUUUUGUGUUCAAGAAAUCAUC-----------------AAGAAGUAUAGUGAGUUUAAACUAACU .(((....((((((((.((((---------....((((.((((....)))).....)))).....(....).))-----------------)))))))))))))((((......)))) ( -15.00) >DroYak_CAF1 3948 117 - 1 AAACAAUUUAUACUUUAUUGACGUUAGCGACUCCUGCCUAGCGACUUCAGAUGAUUUGUGCAUAAGAAAUCAUCUAAUCCCCAAUCUAUUUAAAAAGUAUCGUGAGU-UAAAUUAAGU ....((((((((((((.((((((((((((.....)).)))))).....(((((((((.(.....).)))))))))..............)))))))))).(....).-)))))).... ( -21.40) >consensus ACACAUUUUAUACUUUAUUGA_________CUCCUACCUAGCGAAUUCAGAUAAUUUGUGUACAAGAAACCAUCU_AUCCC_AA__UAUUUAAGAAGUAACGUGGGUUUCA_AUAA__ ..........((((((........................(((((((.....))))))).....(((.....)))..................))))))................... ( -2.62 = -3.18 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:50 2006