| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,218,326 – 10,218,422 |
| Length | 96 |
| Max. P | 0.955536 |

| Location | 10,218,326 – 10,218,422 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 70.29 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -15.55 |
| Energy contribution | -15.80 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.955536 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
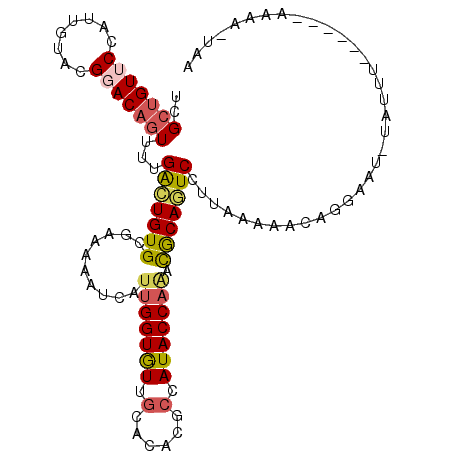
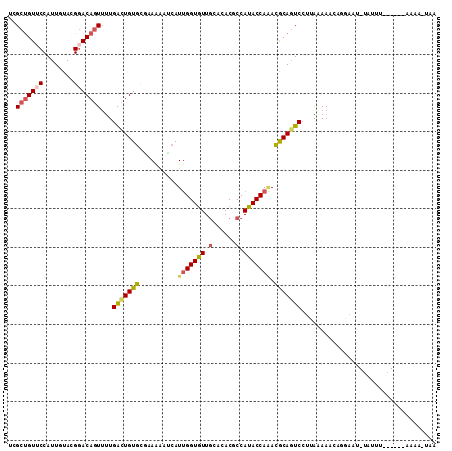
>2L_DroMel_CAF1 10218326 96 + 22407834 UCGCUGUUCCAUUGUACGGACAGUUUUGGCUGUGCGAAAAGUCAUUGGUGUUGCACACGCCAUACCAAACGCAGUCCUUAAAAAGUUGC--------------AAAA-UAA ..(((((((........)))))))...(((((((.((....)).(((((((.((....)).))))))).))))))).............--------------....-... ( -24.90) >DroVir_CAF1 66479 101 + 1 UUGGUGUACCAUUAUAGGAACAAUAUUGAUUGUGAUAAGAAGAAUCGGUAUUACAAACUCCAUACCAGAUACAAUCCUGGAAAAUAUCAAU-UAUUUU--------A-UUA .(((.((.((......))...((((((((((.(.......).))))))))))....)).)))..((((........))))((((((.....-))))))--------.-... ( -18.70) >DroSim_CAF1 3785 96 + 1 UCGCUGUUCCAUUGUACGGACAGUUUUGGCUGUGCGAAUACUCAUUGGUGUUGCACACGCCAUACCAAACGCAGUCCUUACAAAGUGGA--------------AAAA-UAA ......((((((((((.((((.(((((((((((((.((((((....))))))))))).))))....))))...)))).))))..)))))--------------)...-... ( -34.20) >DroEre_CAF1 3269 111 + 1 UCGCUGUUCCAUUGUACGGACAGUUUUGACUGUGACCAUUUUCAUUGGUGUUGCACACUCCAUACCAAACGCAGUCCUUAAAAACAGGAAUAUUUUUAAUAGUAAAAAACC ....((((((..(((....)))(((((((((((((......)).(((((((.(......).)))))))..))))))....))))).))))))((((((....))))))... ( -20.80) >DroYak_CAF1 3874 110 + 1 UCGCUGUUCCAUUAUACGGACAGUUUUGACUGUGCAUAAUAUCAUUGGUGUUGCACACGCCAUACCAAACGCAGUCCUUAGAAACAGAAAUAUAUUUUAUAGUAAAA-GCA ..(((((((........)))).((((((((((((..........(((((((.((....)).))))))).)))))))....))))).....................)-)). ( -24.00) >DroAna_CAF1 3731 108 + 1 UUGCCGUGCCAUUGUAGGGACAGUUCUGGCUGUGUUGAAAGGCAUUGGUGUUGCAAACUCCAUACCACACACAAUCCUUGAAAAAU--AAUAUACAGAAUAAUCAAA-AUU .((((.(((....))).)).))((((((....((((..((((.((((((((.(...........).)))).))))))))....)))--).....)))))).......-... ( -22.70) >consensus UCGCUGUUCCAUUGUACGGACAGUUUUGACUGUGCGAAAAAUCAUUGGUGUUGCACACGCCAUACCAAACGCAGUCCUUAAAAACAGGAAU_UAUUU______AAAA_UAA ..(((((((........)))))))...(((((((..........(((((((.(......).))))))).)))))))................................... (-15.55 = -15.80 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:54 2006