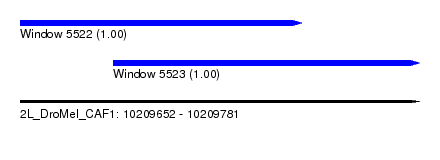
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,209,652 – 10,209,781 |
| Length | 129 |
| Max. P | 0.999963 |

| Location | 10,209,652 – 10,209,743 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 92.42 |
| Mean single sequence MFE | -24.26 |
| Consensus MFE | -22.64 |
| Energy contribution | -22.88 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.93 |
| SVM RNA-class probability | 0.999963 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
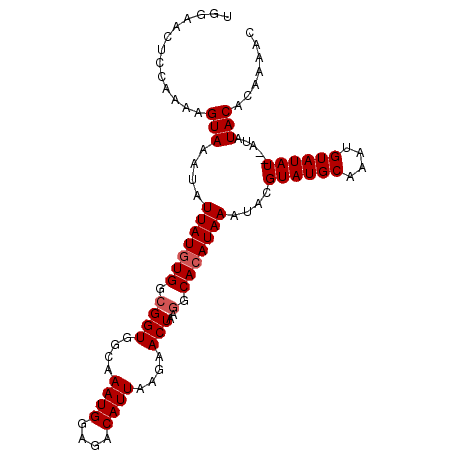
>2L_DroMel_CAF1 10209652 91 + 22407834 GACGCAUCAAUGCCAUUUUAGCCACUUAGAUGGAACUCGAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCA ...((.....((((((....(((((...((((..(((.....)))...))))..))))).))))))((((....))))..........)). ( -25.30) >DroSec_CAF1 2796 91 + 1 AACGCUUCAAUGCCAUUUUAGCCACUUAGAUGGAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAAGCA ...((((...((((((....(((((...((((..(((.....)))...))))..))))).))))))((((....))))........)))). ( -26.50) >DroSim_CAF1 2530 91 + 1 AACGCUUCAAUGCCAUUUUAGCCACUUAGAUGGAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCA ...((((...((((((....(((((...((((..(((.....)))...))))..))))).))))))((((....))))........)))). ( -26.20) >DroEre_CAF1 2708 91 + 1 GGCGCUGUAAUGCCAUUUUAGCCAGUUAGAUAAAACUCCAAAAGUAAAUAUUAUAUGGCGGUGGCAAAUGAAGACAUUAAGAACUAAGGCA ...(((....((((((....((((....((((..(((.....)))...))))...)))).))))))((((....)))).........))). ( -18.20) >DroYak_CAF1 2564 91 + 1 GCAGCUGUAAGGCCAUUUUAGCCACUUAGAUAAAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCA ...(((...(((((((....(((((...((((..(((.....)))...))))..))))).))))).((((....)))).....))..))). ( -25.10) >consensus GACGCUUCAAUGCCAUUUUAGCCACUUAGAUGGAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCA ...(((....((((((....(((((...((((..(((.....)))...))))..))))).))))))((((....)))).........))). (-22.64 = -22.88 + 0.24)


| Location | 10,209,682 – 10,209,781 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 96.39 |
| Mean single sequence MFE | -19.00 |
| Consensus MFE | -16.68 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10209682 99 + 22407834 UGGAACUCGAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCACAUAAAUACGUAUGCAAAUGUAUAU--AUAUACACAAAAC .............(((....(((((((.((((....((((....))))....)))..).)))))))....((((((....))))))--...)))....... ( -20.10) >DroSec_CAF1 2826 99 + 1 UGGAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAAGCACAUAAAUACGUAUGCAAAUGUAUAU--AUAUACACAAAAC (((....)))...(((....(((((((.(..(((..((((....)))).....)))..))))))))....((((((....))))))--...)))....... ( -20.30) >DroSim_CAF1 2560 99 + 1 UGGAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCACAUAAAUACGUAUGCAAAUGUAUAU--AUAUACACAAAAC (((....)))...(((....(((((((.((((....((((....))))....)))..).)))))))....((((((....))))))--...)))....... ( -22.30) >DroEre_CAF1 2738 99 + 1 UAAAACUCCAAAAGUAAAUAUUAUAUGGCGGUGGCAAAUGAAGACAUUAAGAACUAAGGCACAUAAAUACGUAUGCAAAUGUAUAU--AUAUACACAAAAC .............(((..((((.((((((..(((..((((....)))).....)))..)).)))))))).((((((....))))))--...)))....... ( -11.30) >DroYak_CAF1 2594 101 + 1 UAAAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCACAUAAAUACGUAUGCAAAUGUAUAUAUAUAUACACAAAAC .............(((....(((((((.((((....((((....))))....)))..).)))))))(((..(((((....)))))..))).)))....... ( -21.00) >consensus UGGAACUCCAAAAGUAAAUAUUAUGUGGCGGUGGCAAAUGGAGACAUUAAGAACUAAGGCACAUAAAUACGUAUGCAAAUGUAUAU__AUAUACACAAAAC .............(((....(((((((.((((....((((....))))....)))..).)))))))....((((((....)))))).....)))....... (-16.68 = -17.08 + 0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:52 2006