| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,193,567 – 10,193,669 |
| Length | 102 |
| Max. P | 0.915478 |

| Location | 10,193,567 – 10,193,669 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.32 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
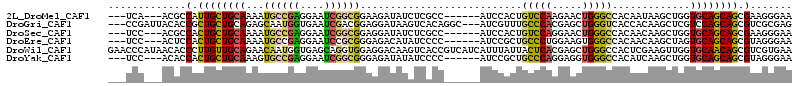
>2L_DroMel_CAF1 10193567 102 + 22407834 UUCCCUUCGCUGCUGCACCAGCUUAUUGUGGCCCAGUUCUUGGACAGUGGAU------GGCGAGAUAUCUUCCGCCGAUUCCUCGGCAUUUUGCAGCAAUGGCGU---UGA--- ....(..((((((((((...((.....)).(((..(((....))).(.((((------((((.((.....)))))))..))).))))....))))))...)))).---.).--- ( -31.50) >DroGri_CAF1 165298 108 + 1 CUCGCGACGCUGCUGGACGAGCUUGUGGUGACCCAGCUCGUGGGCAAACGAU---GCCUGUGACUUAUCCUCCGUCGAUUCACCAUUGCUCUGCAGCAGCGGUGUAAUCGG--- ...(((.((((((((.(.((((..(((((((..(.(((((..((((.....)---)))..)))..........)).)..))))))).))))).)))))))).)))......--- ( -44.50) >DroSec_CAF1 107225 102 + 1 UUCCCUUCGCUGCUGCACCAGCUUGUUGUGGCCCAGUUCCUGGACAGUGGAU------GGCGAGAUAUCCUCCGCCGAUUCCUCGGCAUUUUGCAGCAGUGGCGU---GGA--- .((((.((((((((((((((..(..((((...((((...))))))))..).)------)).(((.....))).(((((....)))))....))))))))))).).---)))--- ( -41.10) >DroEre_CAF1 108858 102 + 1 UUCCCUACGCUGCUGCACUAGCUUGUUGUGGCCCACUUCCAGGGCAGCGGAU------GGGGAUAUGUCUCCCGCGGAUUCCUCGGCAUUUUGCAGCAGUGGAGU---GGA--- .(((((.((((((((((......(((((.(((((.......)))).((((..------.((((....))))))))......).)))))...)))))))))).)).---)))--- ( -42.40) >DroWil_CAF1 196547 114 + 1 UUCACGACGCUGUUGCACCAACUUCGAGUGGCCCAGCUCGUGAGUAAUAAAUGAUGACGGUGACUUGUCCUCCACCUGCUCACCAUUGUUCUGCAACAAGGGUGUUAUGGGUUC ((((..(((((((((((..(((...((((......))))(((((((......((..(.(....))..)).......)))))))....))).))))))...)))))..))))... ( -32.22) >DroYak_CAF1 109090 102 + 1 UUCCCUACGCUGCUGCACCAGCUUGAUGUGGCCCACCUCCUGGGCAGCGGAU------GGGGAUAUAUCUCCCGCCGAUUCCUCGGCACUUUGCAGCAGUGGUGU---GGA--- ....((((((..((((..............(((((.....))))).(((((.------.((((......))))(((((....)))))..))))).))))..))))---)).--- ( -46.20) >consensus UUCCCUACGCUGCUGCACCAGCUUGUUGUGGCCCAGCUCCUGGGCAGUGGAU______GGCGACAUAUCCUCCGCCGAUUCCUCGGCAUUUUGCAGCAGUGGUGU___GGA___ .......((((((((((...(((.......(((((.....))))).............(((((...(((.......)))))))))))....))))))))))............. (-21.84 = -22.32 + 0.48)



| Location | 10,193,567 – 10,193,669 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 71.75 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -22.62 |
| Energy contribution | -22.02 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915478 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
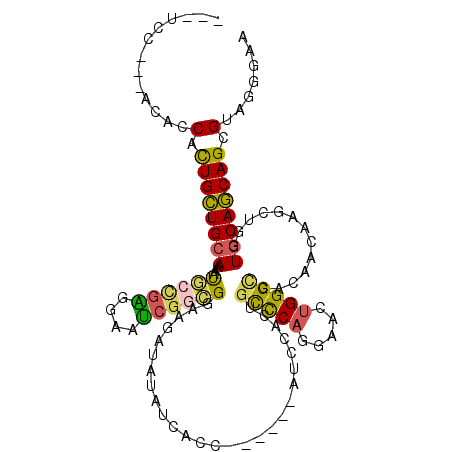
>2L_DroMel_CAF1 10193567 102 - 22407834 ---UCA---ACGCCAUUGCUGCAAAAUGCCGAGGAAUCGGCGGAAGAUAUCUCGCC------AUCCACUGUCCAAGAACUGGGCCACAAUAAGCUGGUGCAGCAGCGAAGGGAA ---...---...((.(((((((....((((((....))))))...........((.------..((((((((((.....))))).......)).))).)).))))))).))... ( -32.21) >DroGri_CAF1 165298 108 - 1 ---CCGAUUACACCGCUGCUGCAGAGCAAUGGUGAAUCGACGGAGGAUAAGUCACAGGC---AUCGUUUGCCCACGAGCUGGGUCACCACAAGCUCGUCCAGCAGCGUCGCGAG ---....(..(..((((((((..((((..((((((..((((.........)))...(((---.((((......))))))))..))))))...))))...))))))))..)..). ( -40.00) >DroSec_CAF1 107225 102 - 1 ---UCC---ACGCCACUGCUGCAAAAUGCCGAGGAAUCGGCGGAGGAUAUCUCGCC------AUCCACUGUCCAGGAACUGGGCCACAACAAGCUGGUGCAGCAGCGAAGGGAA ---(((---.(..(.((((((((....(((((....)))))(((((........))------.)))...((((((...)))))).............)))))))).)..)))). ( -38.30) >DroEre_CAF1 108858 102 - 1 ---UCC---ACUCCACUGCUGCAAAAUGCCGAGGAAUCCGCGGGAGACAUAUCCCC------AUCCGCUGCCCUGGAAGUGGGCCACAACAAGCUAGUGCAGCAGCGUAGGGAA ---(((---.((.(.((((((((....((...(((....(.((((......)))))------.)))...((((.......))))........))...)))))))).).))))). ( -37.10) >DroWil_CAF1 196547 114 - 1 GAACCCAUAACACCCUUGUUGCAGAACAAUGGUGAGCAGGUGGAGGACAAGUCACCGUCAUCAUUUAUUACUCACGAGCUGGGCCACUCGAAGUUGGUGCAACAGCGUCGUGAA ..........((((.(((((....))))).))))....((((..((........))..)))).........((((((((((.((((........))))....)))).)))))). ( -33.70) >DroYak_CAF1 109090 102 - 1 ---UCC---ACACCACUGCUGCAAAGUGCCGAGGAAUCGGCGGGAGAUAUAUCCCC------AUCCGCUGCCCAGGAGGUGGGCCACAUCAAGCUGGUGCAGCAGCGUAGGGAA ---...---...((.((((.((.....(((((....)))))((((......)))).------....(((((((((..((((.....))))...)))).))))).)))))))).. ( -42.90) >consensus ___UCC___ACACCACUGCUGCAAAAUGCCGAGGAAUCGGCGGAAGAUAUAUCACC______AUCCACUGCCCAGGAACUGGGCCACAACAAGCUGGUGCAGCAGCGUAGGGAA .............(.((((((((...((((((....))))))...........................(((((.....))))).............)))))))).)....... (-22.62 = -22.02 + -0.60)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:44 2006