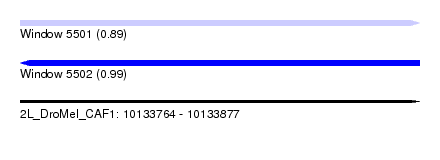
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,133,764 – 10,133,877 |
| Length | 113 |
| Max. P | 0.988782 |

| Location | 10,133,764 – 10,133,877 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 70.35 |
| Mean single sequence MFE | -28.55 |
| Consensus MFE | -10.08 |
| Energy contribution | -12.68 |
| Covariance contribution | 2.60 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.887462 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10133764 113 + 22407834 U-UUCUGGCUGGGACAGCAACUUCUUGCUGGCUUUUGGCCAAUUGUUUAUGGCUUAUUGUUUCAGCUCUCCGGCUUUUUCGCCGGCAACUUGGAAUCGCUUUUGCAAAAGUUUC .-...((((..(((((((((....))))))..)))..)))).((((....(((.....(((((((....(((((......)))))....))))))).)))...))))....... ( -33.80) >DroSec_CAF1 60092 113 + 1 U-UUCUGGCUGGGACAGCAACUGCUCGCUGGCUUUUGGCCAAUUGUUUAUGGCUUAUUGUUUCAGCUCUCCGGCUUUUCCGCCGGCAACUUGGAAUCGCUUUUCCAAAACUUUC .-....(((((..(((((((..(((....)))..)))((((........))))...))))..)))))..(((((......)))))....((((((......))))))....... ( -36.40) >DroSim_CAF1 63531 113 + 1 U-UUCUGUCUGGGACAGCAACUGCUCGCUGGCUUUUGGCCAAUUGUUUAUGGCUUAUUGUUUCAGCUCUCCGGCUUUUUCGCCGGCAACUUGGAAUCGCUUUUCCAAAACUUUC .-....(.(((..(((((((..(((....)))..)))((((........))))...))))..))))...(((((......)))))....((((((......))))))....... ( -31.90) >DroEre_CAF1 58695 111 + 1 U-UUCUGGCUGGGACAGCAACUCCUUGCUGGCUUUUGGCCAAUUGUUUAUGGCUUAUUGUUUCAGCUCUCCGGCUUUUUCGCCGGCAACUUGGAAUCGCUUUU-C-AAAGUUUC .-....(((((..(((((((....))))).......(((((........)))))....))..)))))..(((((......))))).(((((.(((......))-)-.))))).. ( -34.10) >DroWil_CAF1 90608 95 + 1 CAGGCCAUAAAAAACACAGACUUCCAUCUGGCUGGCUCCCAAUUGUUUUUGGUUUU-UGUU------GGUU--UUUUUU--------UUUUGGUGUAUCCUGC-C-AAAGUUAC (((((((.((((((..((((......))))((..((..((((......))))....-.)).------.)).--..))))--------)).))).....)))).-.-........ ( -18.50) >DroAna_CAF1 64378 86 + 1 -------------AAAACUCCUACUUGCUGGCUUUUGGCCAAUUGUUUAUGCCUUAUUGUUUCUGCUCUUUGGCUUUUGCAGCG---------AA-----UUUCU-AAAGUUUC -------------...........((((((.(....((((((..((....((......))....))...))))))...))))))---------).-----.....-........ ( -16.60) >consensus U_UUCUGGCUGGGACAGCAACUGCUUGCUGGCUUUUGGCCAAUUGUUUAUGGCUUAUUGUUUCAGCUCUCCGGCUUUUUCGCCGGCAACUUGGAAUCGCUUUUCC_AAAGUUUC ...((((.((((((((((........))((((.....))))................))))))))....(((((......))))).....)))).................... (-10.08 = -12.68 + 2.60)

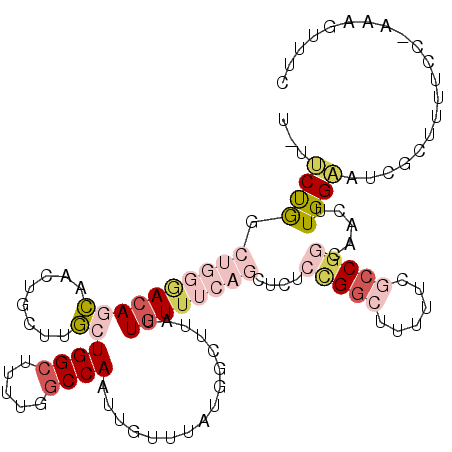
| Location | 10,133,764 – 10,133,877 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 70.35 |
| Mean single sequence MFE | -27.83 |
| Consensus MFE | -10.68 |
| Energy contribution | -12.85 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.38 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988782 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10133764 113 - 22407834 GAAACUUUUGCAAAAGCGAUUCCAAGUUGCCGGCGAAAAAGCCGGAGAGCUGAAACAAUAAGCCAUAAACAAUUGGCCAAAAGCCAGCAAGAAGUUGCUGUCCCAGCCAGAA-A ..(((((((((....))))....))))).(((((......))))).(.((((.........((((........)))).......((((((....))))))...)))))....-. ( -31.70) >DroSec_CAF1 60092 113 - 1 GAAAGUUUUGGAAAAGCGAUUCCAAGUUGCCGGCGGAAAAGCCGGAGAGCUGAAACAAUAAGCCAUAAACAAUUGGCCAAAAGCCAGCGAGCAGUUGCUGUCCCAGCCAGAA-A ......(((((((......)))))))...(((((......))))).(.((((.........((((........)))).......((((((....))))))...)))))....-. ( -32.80) >DroSim_CAF1 63531 113 - 1 GAAAGUUUUGGAAAAGCGAUUCCAAGUUGCCGGCGAAAAAGCCGGAGAGCUGAAACAAUAAGCCAUAAACAAUUGGCCAAAAGCCAGCGAGCAGUUGCUGUCCCAGACAGAA-A ....((((.(((..(((((((.(......(((((......)))))...((((.........((((........)))).......))))..).))))))).))).))))....-. ( -34.09) >DroEre_CAF1 58695 111 - 1 GAAACUUU-G-AAAAGCGAUUCCAAGUUGCCGGCGAAAAAGCCGGAGAGCUGAAACAAUAAGCCAUAAACAAUUGGCCAAAAGCCAGCAAGGAGUUGCUGUCCCAGCCAGAA-A ......((-(-...(((((((((......(((((......)))))...((((.........((((........)))).......))))..)))))))))....)))......-. ( -34.09) >DroWil_CAF1 90608 95 - 1 GUAACUUU-G-GCAGGAUACACCAAAA--------AAAAAA--AACC------AACA-AAAACCAAAAACAAUUGGGAGCCAGCCAGAUGGAAGUCUGUGUUUUUUAUGGCCUG .......(-(-((.((.....))....--------......--....------....-....((((......))))..))))((((...(((((......)))))..))))... ( -16.50) >DroAna_CAF1 64378 86 - 1 GAAACUUU-AGAAA-----UU---------CGCUGCAAAAGCCAAAGAGCAGAAACAAUAAGGCAUAAACAAUUGGCCAAAAGCCAGCAAGUAGGAGUUUU------------- ........-.((((-----((---------(.((((....(((..................)))........(((((.....)))))...)))))))))))------------- ( -17.77) >consensus GAAACUUU_GGAAAAGCGAUUCCAAGUUGCCGGCGAAAAAGCCGGAGAGCUGAAACAAUAAGCCAUAAACAAUUGGCCAAAAGCCAGCAAGAAGUUGCUGUCCCAGCCAGAA_A ..............(((((((.(......(((((......)))))...........................(((((.....)))))...).)))))))............... (-10.68 = -12.85 + 2.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:30 2006