
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,112,962 – 10,113,089 |
| Length | 127 |
| Max. P | 0.993507 |

| Location | 10,112,962 – 10,113,064 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -28.35 |
| Consensus MFE | -17.68 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.965736 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
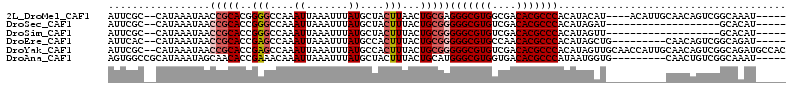
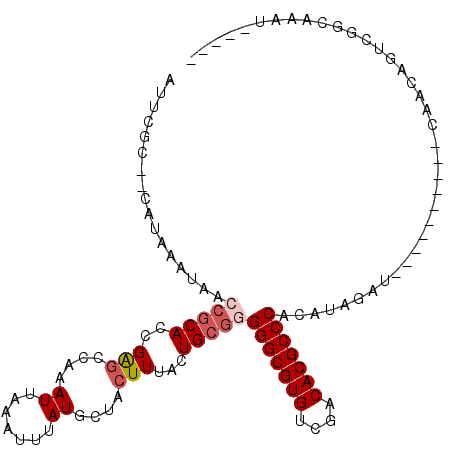
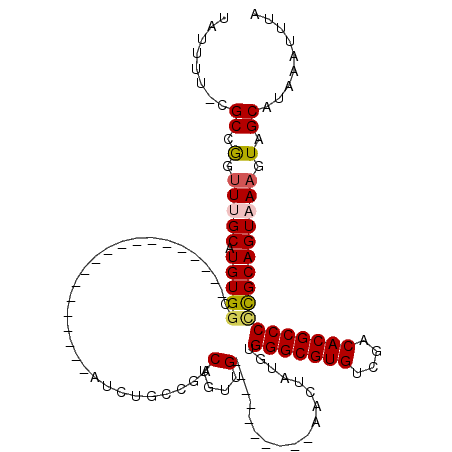
>2L_DroMel_CAF1 10112962 102 + 22407834 AUUCGC--CAUAAAUAACCGCACGGGGCCAAAUUAAAUUUAUGCUACUUAACUGCGAGGGCGUGGCGACACGCCCACAUACAU----ACAUUGCAACAGUCGGCAAAU----- ....((--(........((....))(((.............(((.........))).(((((((....)))))))........----...........))))))....----- ( -27.60) >DroSec_CAF1 39250 87 + 1 AUUCGC--CAUAAAUAACCGCACCGGGCCAAAUUAAAUUUAUGCUACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGAU-------------------GCACAU----- ....((--.........(((((...(((..............))).......)))))(((((((....)))))))........-------------------))....----- ( -23.94) >DroSim_CAF1 42710 87 + 1 AUUCGC--CAUAAAUAACCGCACCGGGCCAAAUUAAAUUUAUGCUACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGUU-------------------GCACAU----- ....((--.((......(((((...(((..............))).......)))))(((((((....))))))).....)).-------------------))....----- ( -23.94) >DroEre_CAF1 37875 97 + 1 AUUCAC--CAUAAAUAACCGCACCGAGCCAAAUUAAAUUUAUGCCACUUUACUGCGGGGGCGUGCCAACACGCCCACAUAGCUG---------CAACAGUCGGCAGAU----- ......--.........(((((..(((....((.......))....)))...)))))(((((((....)))))))......(((---------(........))))..----- ( -25.40) >DroYak_CAF1 39493 111 + 1 AUUCGC--CAUAAAUAACCGCACCGAGCCAAAUUAAAUUUAUGCCACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGUUGCAACCAUUGCAACAGUCGGCAGAUGCCAC ....((--(........(((((..(((....((.......))....)))...)))))(((((((....))))))).....((((((.....))))))....)))......... ( -34.50) >DroAna_CAF1 42538 99 + 1 AGUGGCCGCAUAAAUAGCAACACCGAAACAAAUUAAAUUUAUGCUACUUUACUGCAUGGGCGUGGUGACACGCCCAUAAUGGUG---------CAACUGUCGGCAAAU----- ....((((.....((((...(((((.................((.........))(((((((((....)))))))))..)))))---------...))))))))....----- ( -34.70) >consensus AUUCGC__CAUAAAUAACCGCACCGAGCCAAAUUAAAUUUAUGCUACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGAU__________CAACAGUCGGCAAAU_____ .................(((((..(((....((.......))....)))...)))))(((((((....)))))))...................................... (-17.68 = -18.77 + 1.08)
| Location | 10,112,962 – 10,113,064 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 78.67 |
| Mean single sequence MFE | -30.57 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.38 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.713719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
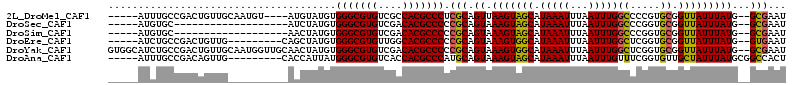
>2L_DroMel_CAF1 10112962 102 - 22407834 -----AUUUGCCGACUGUUGCAAUGU----AUGUAUGUGGGCGUGUCGCCACGCCCUCGCAGUUAAGUAGCAUAAAUUUAAUUUGGCCCCGUGCGGUUAUUUAUG--GCGAAU -----(((((((((((((((((....----.)))).(.(((((((....))))))).)))))))(((((((.(((((...)))))((.....)).)))))))..)--)))))) ( -32.10) >DroSec_CAF1 39250 87 - 1 -----AUGUGC-------------------AUCUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUAGCAUAAAUUUAAUUUGGCCCGGUGCGGUUAUUUAUG--GCGAAU -----...(((-------------------........(((((((....)))))))(((((.(......((.(((((...)))))))..).))))).........--)))... ( -23.10) >DroSim_CAF1 42710 87 - 1 -----AUGUGC-------------------AACUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUAGCAUAAAUUUAAUUUGGCCCGGUGCGGUUAUUUAUG--GCGAAU -----.(((((-------------------.(((.((((((((((....)))))))..)))....))).))))).........(((((......)))))......--...... ( -25.50) >DroEre_CAF1 37875 97 - 1 -----AUCUGCCGACUGUUG---------CAGCUAUGUGGGCGUGUUGGCACGCCCCCGCAGUAAAGUGGCAUAAAUUUAAUUUGGCUCGGUGCGGUUAUUUAUG--GUGAAU -----....((((((((...---------.(((((...(((((((....)))))))((((......)))).............))))))))).))))........--...... ( -30.60) >DroYak_CAF1 39493 111 - 1 GUGGCAUCUGCCGACUGUUGCAAUGGUUGCAACUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUGGCAUAAAUUUAAUUUGGCUCGGUGCGGUUAUUUAUG--GCGAAU ...(((((.(((((..((((((.....))))))((((.(((((((....)))))))((((......))))))))........)))))..))))).((((....))--)).... ( -39.30) >DroAna_CAF1 42538 99 - 1 -----AUUUGCCGACAGUUG---------CACCAUUAUGGGCGUGUCACCACGCCCAUGCAGUAAAGUAGCAUAAAUUUAAUUUGUUUCGGUGUUGCUAUUUAUGCGGCCACU -----....((((..(((.(---------((((..((((((((((....)))))))))).......(.((((...........)))).)))))).))).......)))).... ( -32.80) >consensus _____AUCUGCCGACUGUUG__________AACUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUAGCAUAAAUUUAAUUUGGCCCGGUGCGGUUAUUUAUG__GCGAAU ......................................(((((((....))))))).(((.((.(((((((.(((((...)))))((.....)).)))))))))...)))... (-19.69 = -19.38 + -0.30)
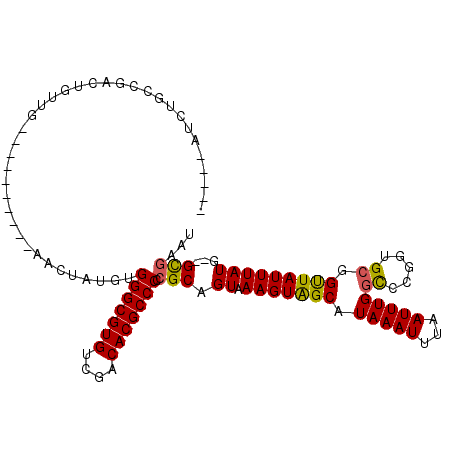

| Location | 10,112,993 – 10,113,089 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -31.18 |
| Consensus MFE | -21.66 |
| Energy contribution | -21.72 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.24 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.993507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10112993 96 + 22407834 UAAAUUUAUGCUACUUAACUGCGAGGGCGUGGCGACACGCCCACAUACAU----ACAUUGCAACAGUCGGCAAAU-------------------GCCACAUGCAAACCGGCGGAAAAUA ..................((((..(((((((....)))))))........----...(((((...((.(((....-------------------))))).)))))....))))...... ( -31.60) >DroSec_CAF1 39281 80 + 1 UAAAUUUAUGCUACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGAU-------------------GCACAU-------------------GCCACAUGCAAACCGGCG-AAAAUA ...((((.((((......(((.(.(((((((....))))))).).))).(-------------------(((...-------------------......))))....))))-.)))). ( -22.60) >DroSim_CAF1 42741 80 + 1 UAAAUUUAUGCUACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGUU-------------------GCACAU-------------------GCCACAUGCAAACCGGCG-AAAAUA ...((((.((((.......((((.(((((((....))))))).....((.-------------------((....-------------------)).)).))))....))))-.)))). ( -23.30) >DroEre_CAF1 37906 90 + 1 UAAAUUUAUGCCACUUUACUGCGGGGGCGUGCCAACACGCCCACAUAGCUG---------CAACAGUCGGCAGAU-------------------GCCACAUGCAAGCCGGCG-AAAAUA ...((((.((((............(((((((....))))))).....((((---------((...((.(((....-------------------))))).))).))).))))-.)))). ( -31.40) >DroYak_CAF1 39524 118 + 1 UAAAUUUAUGCCACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGUUGCAACCAUUGCAACAGUCGGCAGAUGCCACACAUACCCACAGAUGCCACAUGCAAACCGGCG-AAAAUA ...((((.((((......(((.(((((((((....))))))).....((((((.....)))))).((.(((....)))))......)).))).(((.....)))....))))-.)))). ( -41.90) >DroAna_CAF1 42571 90 + 1 UAAAUUUAUGCUACUUUACUGCAUGGGCGUGGUGACACGCCCAUAAUGGUG---------CAACUGUCGGCAAAU-------------------GCCACAUGCGAACUGGCA-GAAAUA ...((((.(((((.((((((..(((((((((....)))))))))...)))(---------((..(((.(((....-------------------)))))))))))).)))))-.)))). ( -36.30) >consensus UAAAUUUAUGCUACUUUACUGCGGGGGCGUGUCGACACGCCCACAUAGAU__________CAACAGUCGGCAAAU___________________GCCACAUGCAAACCGGCG_AAAAUA ....(((.((((.......((((.(((((((....)))))))..........................(((.......................)))...))))....)))).)))... (-21.66 = -21.72 + 0.06)
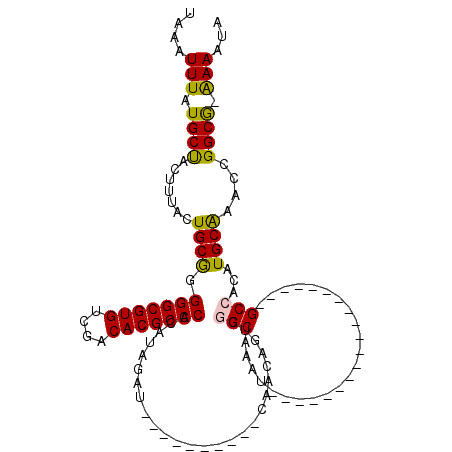
| Location | 10,112,993 – 10,113,089 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 75.71 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -20.49 |
| Energy contribution | -20.85 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.63 |
| SVM decision value | 2.21 |
| SVM RNA-class probability | 0.990396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10112993 96 - 22407834 UAUUUUCCGCCGGUUUGCAUGUGGC-------------------AUUUGCCGACUGUUGCAAUGU----AUGUAUGUGGGCGUGUCGCCACGCCCUCGCAGUUAAGUAGCAUAAAUUUA ........((.(((.(((.....))-------------------)...)))((((((((((....----.)))).(.(((((((....))))))).))))))).....))......... ( -28.90) >DroSec_CAF1 39281 80 - 1 UAUUUU-CGCCGGUUUGCAUGUGGC-------------------AUGUGC-------------------AUCUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUAGCAUAAAUUUA ......-.((.(.(((((.((((((-------------------(((...-------------------...)))).(((((((....))))))))))))))))).).))......... ( -27.40) >DroSim_CAF1 42741 80 - 1 UAUUUU-CGCCGGUUUGCAUGUGGC-------------------AUGUGC-------------------AACUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUAGCAUAAAUUUA ......-.((.(.(((((.((((((-------------------(((...-------------------...)))).(((((((....))))))))))))))))).).))......... ( -27.40) >DroEre_CAF1 37906 90 - 1 UAUUUU-CGCCGGCUUGCAUGUGGC-------------------AUCUGCCGACUGUUG---------CAGCUAUGUGGGCGUGUUGGCACGCCCCCGCAGUAAAGUGGCAUAAAUUUA ......-.(((((((.(((.(((((-------------------....))).))...))---------))))).((((((((((....)))))))..))).......)))......... ( -34.30) >DroYak_CAF1 39524 118 - 1 UAUUUU-CGCCGGUUUGCAUGUGGCAUCUGUGGGUAUGUGUGGCAUCUGCCGACUGUUGCAAUGGUUGCAACUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUGGCAUAAAUUUA ......-.((((.(((((.....))..(((((((..((((((((((..(((.((.((((((.....))))))...)).)))))))).)))))..))))))).))).))))......... ( -48.20) >DroAna_CAF1 42571 90 - 1 UAUUUC-UGCCAGUUCGCAUGUGGC-------------------AUUUGCCGACAGUUG---------CACCAUUAUGGGCGUGUCACCACGCCCAUGCAGUAAAGUAGCAUAAAUUUA ((((((-(((......(((((((((-------------------....))).)))..))---------)......(((((((((....)))))))))))))..)))))........... ( -29.90) >consensus UAUUUU_CGCCGGUUUGCAUGUGGC___________________AUCUGCCGACUGUUG__________AACUAUGUGGGCGUGUCGACACGCCCCCGCAGUAAAGUAGCAUAAAUUUA ........((.(.(((((.(((((.............................(....)..................(((((((....))))))))))))))))).).))......... (-20.49 = -20.85 + 0.36)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:25 2006