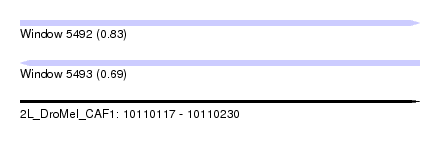
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,110,117 – 10,110,230 |
| Length | 113 |
| Max. P | 0.826743 |

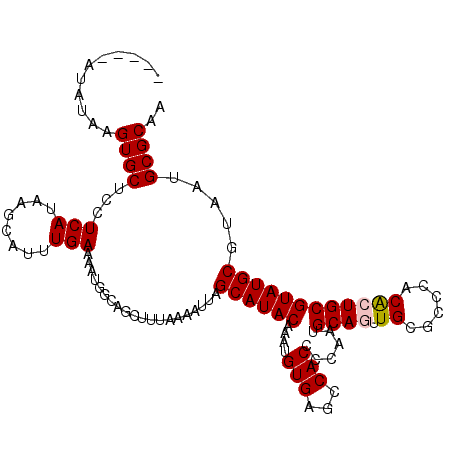
| Location | 10,110,117 – 10,110,230 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -31.32 |
| Consensus MFE | -23.55 |
| Energy contribution | -24.26 |
| Covariance contribution | 0.71 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.826743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10110117 113 + 22407834 UUGCGCAUUACGCAUACGCAGUGUGGGCGCAAUUGCAUUGGGGUGGCUCACAUUUGUAUGCAAAUUUUAAAGCUGCCAUUUUCAAAUGCUUAUGAGGAGCACUGAUGUUUUUU ((((((.((((((.......))))))))))))..((((..(((..(((...(((((....))))).....)))..)).........(((((.....))))))..))))..... ( -36.00) >DroSec_CAF1 36415 108 + 1 UUGCGCAUUACGCAUACGCAGUGUGGGCGCAACUGCAUUGGGGUGGCUCACAUUUGUAUGCUAAUUUUAAAGCAGUCAUUUUCAAAUGCUUAUGAGAAGCACUUAUAU----- .(((.......(((((((.(((((((((((..(......)..)).))))))))))))))))...((((((((((............))))).))))).))).......----- ( -32.00) >DroSim_CAF1 39876 108 + 1 UUGCGCAUUACGCAUACGCAGUGUGGGCGCAAAUGCAUUGGGGUGGCUCACAUUUGUAUGCUAAUUUUAAAGCAGCCAUUCUCAAAUGCUUAUGAGAAGCACUUAUAU----- ((((((.((((((.......))))))))))))..(((((((((((((..((....)).((((........))))))))))))..))))).((((((.....)))))).----- ( -34.60) >DroEre_CAF1 35058 91 + 1 UUGCGCAUUACGCAUACGCAGCGUGGGUGCAUCUGCAUUGGGGUGGCUCACAUUCGUAUGCAAAUUUUAAAGCUUCCAUUU-----------------GCACUCCCAC----- ...........(((((((.((.((((((.(((((......))))))))))).)))))))))..........((........-----------------))........----- ( -24.40) >DroYak_CAF1 36574 108 + 1 UUGCGCAUUACGCAUACGCAGUGUGGGUGCAGCUGCAUUGGGGUGCUUCACAUUUGUAUGCUAAUUUUAAAGCUUCCACUUUCAAAUGCUCAUGAGGAGCACUUACAU----- ..((..((((.(((((((.((((((((.(((.((......)).))))))))))))))))))))))......)).............(((((.....))))).......----- ( -29.60) >consensus UUGCGCAUUACGCAUACGCAGUGUGGGCGCAACUGCAUUGGGGUGGCUCACAUUUGUAUGCUAAUUUUAAAGCUGCCAUUUUCAAAUGCUUAUGAGAAGCACUUAUAU_____ .(((.......(((((((.(((((((((.((.((......)).)))))))))))))))))).................((((((........)))))))))............ (-23.55 = -24.26 + 0.71)

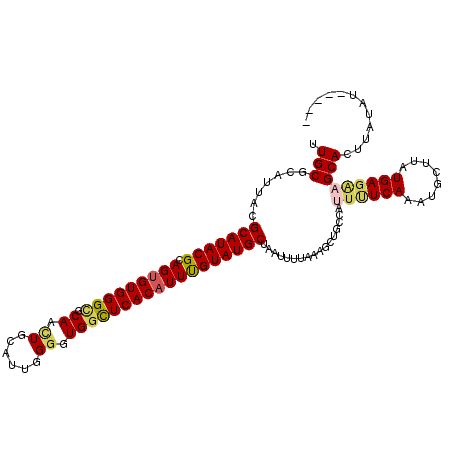
| Location | 10,110,117 – 10,110,230 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.09 |
| Mean single sequence MFE | -28.34 |
| Consensus MFE | -16.69 |
| Energy contribution | -17.08 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10110117 113 - 22407834 AAAAAACAUCAGUGCUCCUCAUAAGCAUUUGAAAAUGGCAGCUUUAAAAUUUGCAUACAAAUGUGAGCCACCCCAAUGCAAUUGCGCCCACACUGCGUAUGCGUAAUGCGCAA ........((((((((.......)))).))))....(((.((((....(((((....)))))..)))).........((....))))).....(((((((.....))))))). ( -25.80) >DroSec_CAF1 36415 108 - 1 -----AUAUAAGUGCUUCUCAUAAGCAUUUGAAAAUGACUGCUUUAAAAUUAGCAUACAAAUGUGAGCCACCCCAAUGCAGUUGCGCCCACACUGCGUAUGCGUAAUGCGCAA -----...(((((((((.....))))))))).....((((((..........((.(((....))).)).........))))))((((.......)))).((((.....)))). ( -26.31) >DroSim_CAF1 39876 108 - 1 -----AUAUAAGUGCUUCUCAUAAGCAUUUGAGAAUGGCUGCUUUAAAAUUAGCAUACAAAUGUGAGCCACCCCAAUGCAUUUGCGCCCACACUGCGUAUGCGUAAUGCGCAA -----...(((((((((((((........))))))((((((((........))).(((....)))))))).......))))))).........(((((((.....))))))). ( -30.20) >DroEre_CAF1 35058 91 - 1 -----GUGGGAGUGC-----------------AAAUGGAAGCUUUAAAAUUUGCAUACGAAUGUGAGCCACCCCAAUGCAGAUGCACCCACGCUGCGUAUGCGUAAUGCGCAA -----(((((.((((-----------------(...((..((((....(((((....)))))..))))..))....)))....)).)))))..(((((((.....))))))). ( -27.80) >DroYak_CAF1 36574 108 - 1 -----AUGUAAGUGCUCCUCAUGAGCAUUUGAAAGUGGAAGCUUUAAAAUUAGCAUACAAAUGUGAAGCACCCCAAUGCAGCUGCACCCACACUGCGUAUGCGUAAUGCGCAA -----...(((((((((.....)))))))))...((((..(((........)))........(((.(((...........))).)))))))..(((((((.....))))))). ( -31.60) >consensus _____AUAUAAGUGCUCCUCAUAAGCAUUUGAAAAUGGCAGCUUUAAAAUUAGCAUACAAAUGUGAGCCACCCCAAUGCAGUUGCGCCCACACUGCGUAUGCGUAAUGCGCAA ...........((((...(((........)))....................((((((....(((...)))......((((.((......)))))))))))).....)))).. (-16.69 = -17.08 + 0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:21 2006