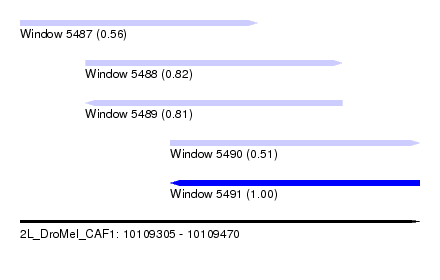
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,109,305 – 10,109,470 |
| Length | 165 |
| Max. P | 0.995351 |

| Location | 10,109,305 – 10,109,403 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.98 |
| Mean single sequence MFE | -27.34 |
| Consensus MFE | -16.04 |
| Energy contribution | -16.44 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.555445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10109305 98 + 22407834 ACACACAC--ACACUUGCAGGCACAGAGAC--UGGAUUCAGGAUUCAGGAAUCCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAU ......((--((....((.((((.(((.((--(((((((.........))))))).)).))))))).))....(((......)))))))............. ( -29.50) >DroSec_CAF1 35647 78 + 1 ACACACAC--AC-------------GAGAC--UGGAUUCAGGAUU-------CCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAU ......((--((-------------((..(--((....)))....-------.((((((((..((.....)).))))))))..))))))............. ( -22.10) >DroSim_CAF1 39098 78 + 1 ACACACAC--AC-------------GAGAC--UGGAUUCAGGAUU-------CCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAU ......((--((-------------((..(--((....)))....-------.((((((((..((.....)).))))))))..))))))............. ( -22.10) >DroEre_CAF1 34274 96 + 1 ACACACAC--AU----GCAGGCAUAGAGACCGGGGAUUCAGGAUUCAGGAUUCCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAU ....((((--((----(((((((.(((.(((.(((((((........))))))).))).)))))))((.((....))))))))).))))............. ( -34.70) >DroYak_CAF1 35752 98 + 1 AUACACACAGAU----ACCGGAUUCAGGAGGCAGGAUUCAGGAUUCAGAAUUCCAGGUGUCUUGCCGACUGUUGACACCUGCGUCGUGUACUUCAAUUAAAU .(((((......----........(((..((((((((.(.(((........)))..).))))))))..)))..(((......))))))))............ ( -28.30) >consensus ACACACAC__AC_____C_GG____GAGAC__UGGAUUCAGGAUUCAG_A_UCCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAU ..((((...........................(((((...))))).......((((((((..((.....)).))))))))....))))............. (-16.04 = -16.44 + 0.40)

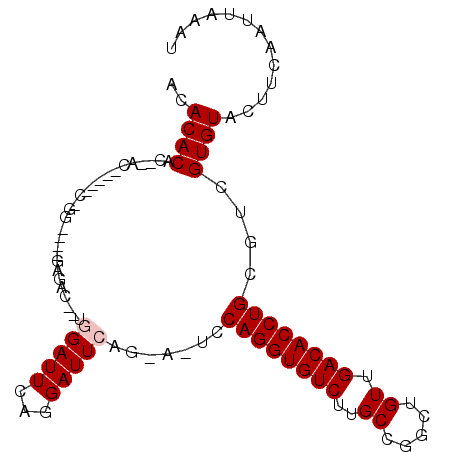
| Location | 10,109,332 – 10,109,438 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -29.72 |
| Consensus MFE | -23.48 |
| Energy contribution | -24.37 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10109332 106 + 22407834 C--UGGAUUCAGGAUUCAGGAAUCCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA-----UAUUCGUGUGCGAACACA (--(((((((.........))))))))((((...((.((.((....)))).)).(((..(((((((((((.....))))))))..(((..-----..))))))..))))))). ( -32.20) >DroSec_CAF1 35661 99 + 1 C--UGGAUUCAGGAUU-------CCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAAA-----UAUUCGUGUGCGAACACA (--((((........)-------))))((((...((.((.((....)))).)).(((..(((((((((((.....))))))))..(((..-----..))))))..))))))). ( -28.30) >DroSim_CAF1 39112 99 + 1 C--UGGAUUCAGGAUU-------CCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA-----UAUUCGUGUGCGAACACA (--((((........)-------))))((((...((.((.((....)))).)).(((..(((((((((((.....))))))))..(((..-----..))))))..))))))). ( -28.30) >DroEre_CAF1 34297 108 + 1 CCGGGGAUUCAGGAUUCAGGAUUCCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA-----UAUUCGUGUGCGAACACA .(((((((((........)))))))(((((((..((.....)).))))))).))(((..(((((((((((.....))))))))..(((..-----..))))))..)))..... ( -31.60) >DroYak_CAF1 35777 108 + 1 GGCAGGAUUCAGGAUUCAGAAUUCCAGGUGUCUUGCCGACUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA-----UAUUCGUGUGCGCACACA (((.((((((........))))))((((((((..((.....)).)))))))).)))((((((((((((((.....))))))))(((((..-----..)))))))))))..... ( -34.20) >DroAna_CAF1 38679 92 + 1 -------------------GUUUCCAGGUGUCUUGCCGGCUGUUGACAUCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUACGACAUACACACAUGCCACU--A -------------------.....((((((((..((.....)).)))))))).(((((((.(((((((((.....))))))))..).)))))))................--. ( -23.70) >consensus C__UGGAUUCAGGAUU___GAUUCCAGGUGUCUUGCCGGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA_____UAUUCGUGUGCGAACACA ........................((((((((..((.....)).))))))))..(((..(((((((((((.....))))))))..(((((.....))))))))..)))..... (-23.48 = -24.37 + 0.89)
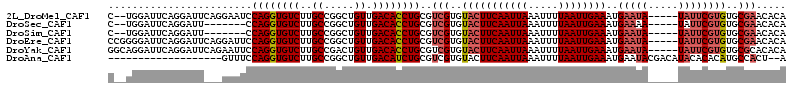
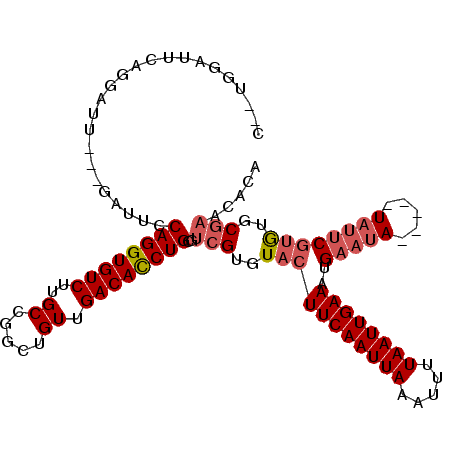
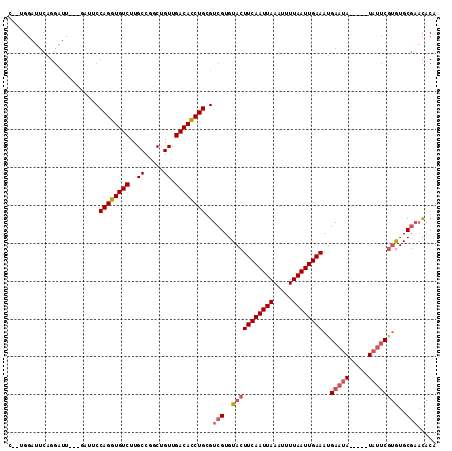
| Location | 10,109,332 – 10,109,438 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.83 |
| Mean single sequence MFE | -27.12 |
| Consensus MFE | -20.94 |
| Energy contribution | -21.97 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.809731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10109332 106 - 22407834 UGUGUUCGCACACGAAUA-----UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCCGGCAAGACACCUGGAUUCCUGAAUCCUGAAUCCA--G ((((((((....))))))-----))...((((((((((.....)))))))))).........(((((((............)))))))((((((..(....).)))))).--. ( -28.90) >DroSec_CAF1 35661 99 - 1 UGUGUUCGCACACGAAUA-----UUUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCCGGCAAGACACCUGG-------AAUCCUGAAUCCA--G .(((((((....))))))-----).(((((((((((((.....))))))))))........((((((((............)))))))))-------))..(((....))--) ( -25.40) >DroSim_CAF1 39112 99 - 1 UGUGUUCGCACACGAAUA-----UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCCGGCAAGACACCUGG-------AAUCCUGAAUCCA--G ((((((((....))))))-----))(((((((((((((.....))))))))))........((((((((............)))))))))-------))..(((....))--) ( -26.80) >DroEre_CAF1 34297 108 - 1 UGUGUUCGCACACGAAUA-----UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCCGGCAAGACACCUGGAAUCCUGAAUCCUGAAUCCCCGG ((((((((....))))))-----))(((((((((((((.....)))))))))).((.((..((((((((............))))))))...)).))......)))....... ( -26.90) >DroYak_CAF1 35777 108 - 1 UGUGUGCGCACACGAAUA-----UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGUCGGCAAGACACCUGGAAUUCUGAAUCCUGAAUCCUGCC .((((((......(((..-----..)))..((((((((.....))))))))))))))...(((((..(((...(((.....)))....(((........))))))..))))). ( -29.10) >DroAna_CAF1 38679 92 - 1 U--AGUGGCAUGUGUGUAUGUCGUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGAUGUCAACAGCCGGCAAGACACCUGGAAAC------------------- .--..((((((.(((((.(((.((((....((((((((.....)))))))))))))))))))).))))))....((((........))))....------------------- ( -25.60) >consensus UGUGUUCGCACACGAAUA_____UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCCGGCAAGACACCUGGAAUC___AAUCCUGAAUCCA__G (((((........(((((.....)))))((((((((((.....)))))))))))))))...((((((((............))))))))........................ (-20.94 = -21.97 + 1.03)

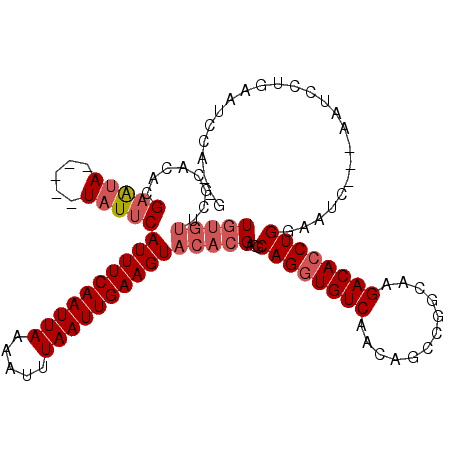
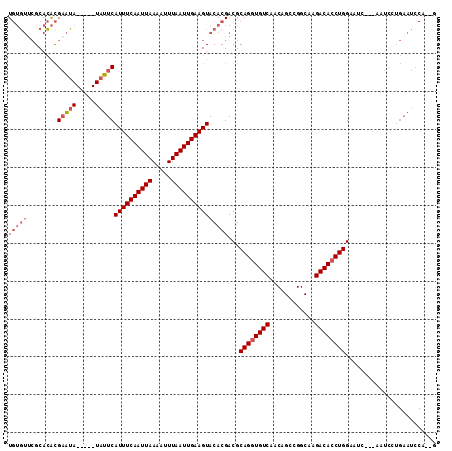
| Location | 10,109,367 – 10,109,470 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -22.78 |
| Consensus MFE | -8.10 |
| Energy contribution | -7.99 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.36 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.511456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10109367 103 + 22407834 GGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA-----UAUUCGUGUGCGAACACAUGCUA-------GUGUGCAUCUUUUUUGUCGCCUCCUUU (((.....((((..((((.((((..((((((((.....)))))))).......-----...(((....)))))))))))..-------))))(((.......))).)))...... ( -24.70) >DroPse_CAF1 33057 82 + 1 GGCUGUUGACAUCUGCG-----UACUUCAAUUAAAAUUUAAUUGAAAUACAUA-------------------CAUAUGUCU-------GUGUACAUCCUUUUUGCCGCCUUC--C (((.((.(((((.((.(-----((.((((((((.....)))))))).)))...-------------------)).)))))(-------(....))........)).)))...--. ( -16.60) >DroSec_CAF1 35689 103 + 1 GGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAAA-----UAUUCGUGUGCGAACACAUGCUA-------GUGUGCAUCUUUUUUGUCGCCUCCUUC (((.....((((..((((.((((..((((((((.....)))))))).......-----...(((....)))))))))))..-------))))(((.......))).)))...... ( -24.70) >DroYak_CAF1 35814 103 + 1 GACUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA-----UAUUCGUGUGCGCACACAUGCUA-------GUGUGCAUCUUUUUUGUCGCCUCCAUC (((....(((......)))..((((((((((((.....))))))))(((((..-----..)))))))))((((((......-------)))))).........)))......... ( -25.00) >DroAna_CAF1 38697 111 + 1 GGCUGUUGACAUCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUACGACAUACACACAUGCCACU--AUACUAUGUAUGUAUGUGCAUCUUUUUUGUGGCCUUC--C (((..(.......((((((((((.(((((((((.....))))))))..).)))))))..(((.((((((....--.......)))))).))))))........)..)))...--. ( -29.06) >DroPer_CAF1 33069 82 + 1 GGCUGUUGACAUCUGCG-----UACUUCAAUUAAAAUUUAAUUGAAAUACAUA-------------------CAUAUGUCU-------GUGUACAUCCUUUUUGCCGCCUUC--C (((.((.(((((.((.(-----((.((((((((.....)))))))).)))...-------------------)).)))))(-------(....))........)).)))...--. ( -16.60) >consensus GGCUGUUGACACCUGCGUCGUGUACUUCAAUUAAAUUUUAAUUGAAAUGAAUA_____UAUUCGUGUGCGAACACAUGCUA_______GUGUGCAUCUUUUUUGUCGCCUCC__C (((......................((((((((.....)))))))).............................((((.............))))..........)))...... ( -8.10 = -7.99 + -0.11)

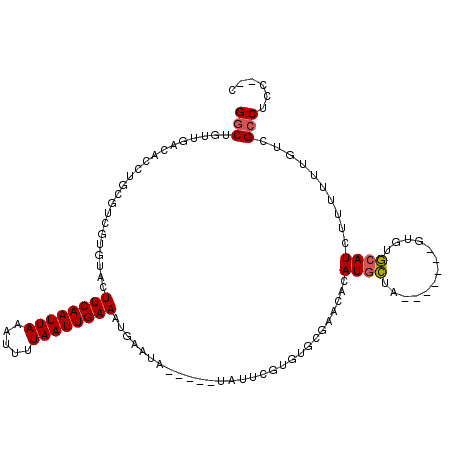
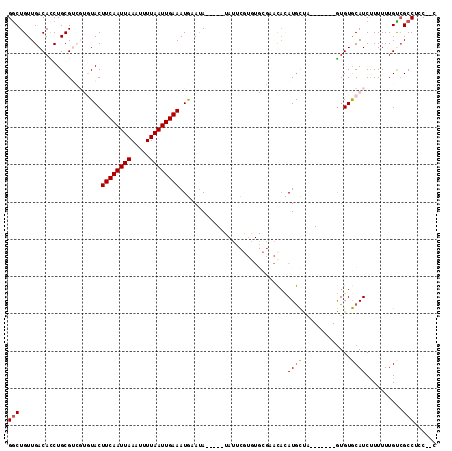
| Location | 10,109,367 – 10,109,470 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.87 |
| Mean single sequence MFE | -24.97 |
| Consensus MFE | -13.13 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.53 |
| SVM decision value | 2.57 |
| SVM RNA-class probability | 0.995351 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10109367 103 - 22407834 AAAGGAGGCGACAAAAAAGAUGCACAC-------UAGCAUGUGUUCGCACACGAAUA-----UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCC ......(((..(......).((.((((-------(.(((((((((((....))))))-----)))..((((((((((.....)))))))))).......)).)))))))...))) ( -25.30) >DroPse_CAF1 33057 82 - 1 G--GAAGGCGGCAAAAAGGAUGUACAC-------AGACAUAUG-------------------UAUGUAUUUCAAUUAAAUUUUAAUUGAAGUA-----CGCAGAUGUCAACAGCC .--...(((.(((.......)))....-------.(((((.((-------------------(..((((((((((((.....)))))))))))-----)))).)))))....))) ( -22.90) >DroSec_CAF1 35689 103 - 1 GAAGGAGGCGACAAAAAAGAUGCACAC-------UAGCAUGUGUUCGCACACGAAUA-----UUUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGCC ......(((..(......).((.((((-------(.((..(((((((....))))))-----)....((((((((((.....)))))))))).......)).)))))))...))) ( -23.10) >DroYak_CAF1 35814 103 - 1 GAUGGAGGCGACAAAAAAGAUGCACAC-------UAGCAUGUGUGCGCACACGAAUA-----UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGGUGUCAACAGUC .........((((........((((((-------......))))))((....(((..-----..)))((((((((((.....)))))))))).......))...))))....... ( -24.50) >DroAna_CAF1 38697 111 - 1 G--GAAGGCCACAAAAAAGAUGCACAUACAUACAUAGUAU--AGUGGCAUGUGUGUAUGUCGUAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGAUGUCAACAGCC .--...(((.........(((((((((((((((((.((..--....)))))))))))))).))))).((((((((((.....))))))))))....((((....))))....))) ( -31.10) >DroPer_CAF1 33069 82 - 1 G--GAAGGCGGCAAAAAGGAUGUACAC-------AGACAUAUG-------------------UAUGUAUUUCAAUUAAAUUUUAAUUGAAGUA-----CGCAGAUGUCAACAGCC .--...(((.(((.......)))....-------.(((((.((-------------------(..((((((((((((.....)))))))))))-----)))).)))))....))) ( -22.90) >consensus G__GAAGGCGACAAAAAAGAUGCACAC_______UAGCAUGUGUUCGCACACGAAUA_____UAUUCAUUUCAAUUAAAAUUUAAUUGAAGUACACGACGCAGAUGUCAACAGCC ......(((((((......((((.............))))...........................((((((((((.....))))))))))............))))....))) (-13.13 = -12.35 + -0.77)

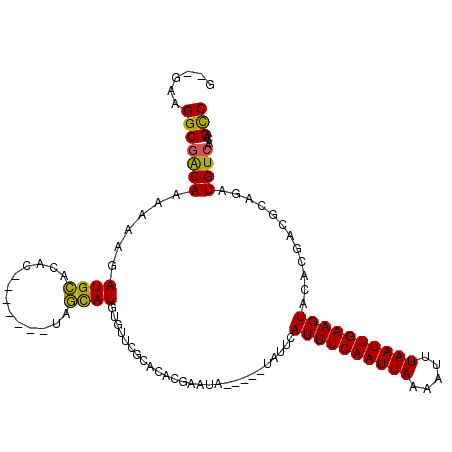
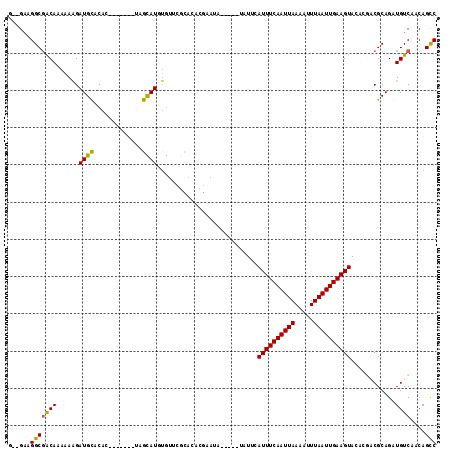
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:53:20 2006