| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,060,704 – 1,060,795 |
| Length | 91 |
| Max. P | 0.982958 |

| Location | 1,060,704 – 1,060,795 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -32.38 |
| Consensus MFE | -27.77 |
| Energy contribution | -27.78 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.982958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
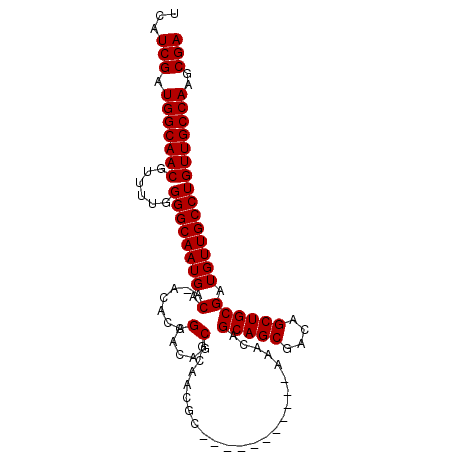
>2L_DroMel_CAF1 1060704 91 + 22407834 UCAUCGUUGGCAACGUUUUGGGGCAAUGACA-ACACAGCAUAACGCAACGC-----------AAACAGCAGCGACAGCUGCGAUGUUGCCUGUUGCCAAGCGA ...(((((((((((......((((((((.(.-.....((.....)).....-----------.....(((((....)))))).)))))))))))))).))))) ( -32.90) >DroSec_CAF1 42942 87 + 1 UCAUCGAUGGCAACGUUUUGGGGCAAUGACAAACACAGCACAGCG-----C-----------AAACAGCAGCGACAGCUGCGAUGUUGCCUGUUGCCAAGCGA ...(((.(((((((......((((((((.(.......((.....)-----)-----------.....(((((....)))))).)))))))))))))))..))) ( -31.20) >DroSim_CAF1 34603 92 + 1 UCAUCGAUGGCAACGUUUUGGGGCAAUGACAAACACAGCACAGCGCAACGC-----------AAACAGCAGCGACAGCUGCGAUGUUGCCUGUUGCCAAGCGA ...(((.(((((((......((((((((.(............(((...)))-----------.....(((((....)))))).)))))))))))))))..))) ( -32.00) >DroEre_CAF1 40021 102 + 1 UCAUCGAUGGCAACGUUUUGGGGCAAUGACA-ACACAGCGCAGCACAACGCAACACAAACAGAAACAGCAGCGACAGCUGCGAUGUUGCCUGUUGCCAAGCGA ...(((.((((((((((.((..((..((...-...))..))..)).)))(((((((.....).....(((((....)))))..))))))..)))))))..))) ( -33.40) >consensus UCAUCGAUGGCAACGUUUUGGGGCAAUGACA_ACACAGCACAGCGCAACGC___________AAACAGCAGCGACAGCUGCGAUGUUGCCUGUUGCCAAGCGA ...(((.(((((((......((((((((.(.......(.....).......................(((((....)))))).)))))))))))))))..))) (-27.77 = -27.78 + 0.00)

| Location | 1,060,704 – 1,060,795 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 88.53 |
| Mean single sequence MFE | -33.73 |
| Consensus MFE | -26.12 |
| Energy contribution | -26.62 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.21 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
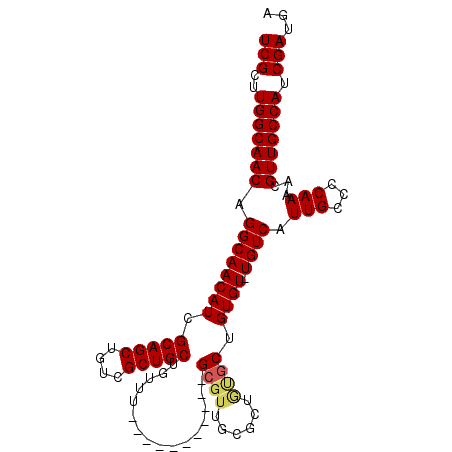
>2L_DroMel_CAF1 1060704 91 - 22407834 UCGCUUGGCAACAGGCAACAUCGCAGCUGUCGCUGCUGUUU-----------GCGUUGCGUUAUGCUGUGU-UGUCAUUGCCCCAAAACGUUGCCAACGAUGA (((.((((((((.(((((((.((((((...((((((.....-----------)))..)))....)))))).-)))...)))).......)))))))))))... ( -34.70) >DroSec_CAF1 42942 87 - 1 UCGCUUGGCAACAGGCAACAUCGCAGCUGUCGCUGCUGUUU-----------G-----CGCUGUGCUGUGUUUGUCAUUGCCCCAAAACGUUGCCAUCGAUGA (((..(((((((.(((((((.((((((.(.(((........-----------)-----)))...))))))..)))...)))).......))))))).)))... ( -30.40) >DroSim_CAF1 34603 92 - 1 UCGCUUGGCAACAGGCAACAUCGCAGCUGUCGCUGCUGUUU-----------GCGUUGCGCUGUGCUGUGUUUGUCAUUGCCCCAAAACGUUGCCAUCGAUGA (((..(((((((.(((((((.((((((.(.((((((.....-----------)))..))))...))))))..)))...)))).......))))))).)))... ( -31.90) >DroEre_CAF1 40021 102 - 1 UCGCUUGGCAACAGGCAACAUCGCAGCUGUCGCUGCUGUUUCUGUUUGUGUUGCGUUGUGCUGCGCUGUGU-UGUCAUUGCCCCAAAACGUUGCCAUCGAUGA (((..(((((((.((((((((((((((...(((.((.............)).)))....))))))..))))-)))).(((...)))...))))))).)))... ( -37.92) >consensus UCGCUUGGCAACAGGCAACAUCGCAGCUGUCGCUGCUGUUU___________GCGUUGCGCUGUGCUGUGU_UGUCAUUGCCCCAAAACGUUGCCAUCGAUGA (((..(((((((.((((((((.(((((....)))))................((((......)))).)))).)))).(((...)))...))))))).)))... (-26.12 = -26.62 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:36 2006