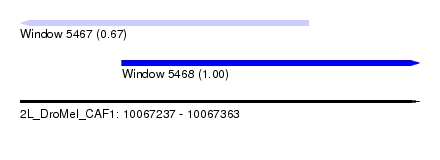
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,067,237 – 10,067,363 |
| Length | 126 |
| Max. P | 0.998112 |

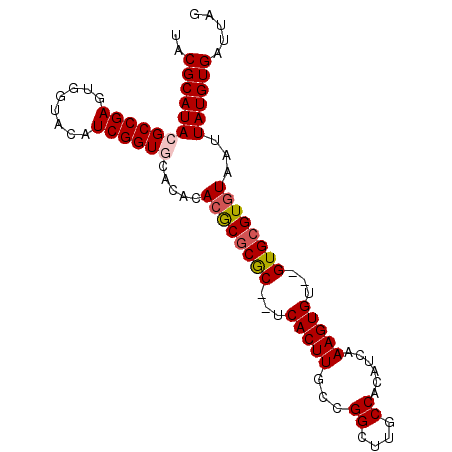
| Location | 10,067,237 – 10,067,328 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 91.07 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -24.36 |
| Energy contribution | -23.68 |
| Covariance contribution | -0.68 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.42 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.672414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10067237 91 - 22407834 GCAAGCCGGCAAGUGAGAGCGCGCUUGUGUGCACCGAUGUACCACUCGGCGUAUGCGUAAUGCGCACAUGGCACAUUCAAAUUCAAACACA----- ....((((....((....))((((.(((((((.((((........)))).)))))))....))))...))))...................----- ( -28.40) >DroSec_CAF1 2421 89 - 1 GGAAGCCGGCAAGUGA--GCGCGCGUGUGUGCACCGAUGUACCACUCGGCGUAUGCGUAAUGCGCACAUGGCACAUUCAAAUUCAAACACA----- ....((((((......--))((((((((((((.((((........)))).))))))...))))))...))))...................----- ( -27.80) >DroSim_CAF1 2437 87 - 1 GGAAGCCGGCAAGUGA--GCGCGCGUGUGUGCACCGAUGUACCACUCGGCGUAUGCGUAAUGCGCACAUGGCACAUUCAAAUUCAAACA------- ....((((((......--))((((((((((((.((((........)))).))))))...))))))...)))).................------- ( -27.80) >DroEre_CAF1 2404 89 - 1 GCAAGCCGGCAAGUGA--GCGCGCGUGUGUGUACCGAUGUGCCACUCGGCGUAUGCGUAAUGCGCACAUGGCACAUUCAAAUUCAAACACA----- ((..((((.....)).--))..))((((.((....(((((((((....((((((.....))))))...)))))))))......)).)))).----- ( -30.40) >DroYak_CAF1 2373 94 - 1 GCCAGCCGGCAAGUGU--GUGUGUGUGUGUAUACCGAUGUGCCACUCGGCGUAUGCGUAAUGCGCACAUGGCACAUUCAAAUUCAAACACAAAGAC (((....)))....((--.(.(((((.((......(((((((((....((((((.....))))))...)))))))))......)).))))).).)) ( -33.00) >consensus GCAAGCCGGCAAGUGA__GCGCGCGUGUGUGCACCGAUGUACCACUCGGCGUAUGCGUAAUGCGCACAUGGCACAUUCAAAUUCAAACACA_____ ....((((((........))((((((((((((.((((........)))).))))))...))))))...))))........................ (-24.36 = -23.68 + -0.68)

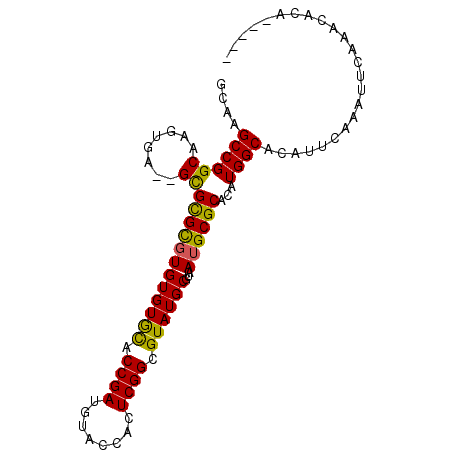
| Location | 10,067,269 – 10,067,363 |
|---|---|
| Length | 94 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 93.38 |
| Mean single sequence MFE | -33.74 |
| Consensus MFE | -28.76 |
| Energy contribution | -29.24 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.51 |
| Structure conservation index | 0.85 |
| SVM decision value | 3.01 |
| SVM RNA-class probability | 0.998112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10067269 94 + 22407834 UACGCAUACGCCGAGUGGUACAUCGGUGCACACAAGCGCGCUCUCACUUGCCGGCUUGCCACAUCAAAGUGU--GUGCGUGUAAUUAUGUGAUUAG (((((((((((.((((((((..(((((((......))))((........)))))..)))))).))...))))--)))))))............... ( -33.50) >DroSec_CAF1 2453 92 + 1 UACGCAUACGCCGAGUGGUACAUCGGUGCACACACGCGCGC--UCACUUGCCGGCUUCCCACAUCAAAGUGU--GUGCGUGUAAUUAUGUGAUUAG ..((((((((((((........)))))).....((((((((--.(((((...((....))......))))).--))))))))...))))))..... ( -32.80) >DroSim_CAF1 2467 92 + 1 UACGCAUACGCCGAGUGGUACAUCGGUGCACACACGCGCGC--UCACUUGCCGGCUUCCCACAUCAAAGUGU--GUGCGUGUAAUUAUGUGAUUAG ..((((((((((((........)))))).....((((((((--.(((((...((....))......))))).--))))))))...))))))..... ( -32.80) >DroEre_CAF1 2436 92 + 1 UACGCAUACGCCGAGUGGCACAUCGGUACACACACGCGCGC--UCACUUGCCGGCUUGCCACAUCAAAGUGU--GUGCGUGUAAUUAUGUGAUUAG ..((((((.(((((........)))))......((((((((--.(((((...((....))......))))).--))))))))...))))))..... ( -32.70) >DroYak_CAF1 2410 94 + 1 UACGCAUACGCCGAGUGGCACAUCGGUAUACACACACACAC--ACACUUGCCGGCUGGCCACAUCAAAGUGUGUGUGCGUGUAAUUAUGUGAUUAG ..((((((.(((((........)))))....((((.(((((--(((((((((....))).......))))))))))).))))...))))))..... ( -36.91) >consensus UACGCAUACGCCGAGUGGUACAUCGGUGCACACACGCGCGC__UCACUUGCCGGCUUGCCACAUCAAAGUGU__GUGCGUGUAAUUAUGUGAUUAG ..((((((((((((........)))))).....((((((((...(((((...((....))......)))))...))))))))...))))))..... (-28.76 = -29.24 + 0.48)

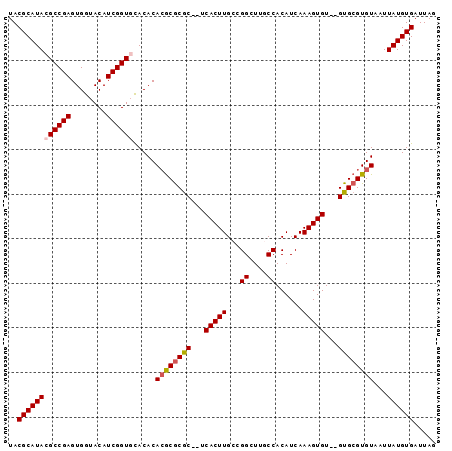
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:57 2006