| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,048,014 – 10,048,120 |
| Length | 106 |
| Max. P | 0.994382 |

| Location | 10,048,014 – 10,048,120 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -29.47 |
| Consensus MFE | -17.47 |
| Energy contribution | -19.30 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10048014 106 + 22407834 UCUAUUUUUUUUUCUAUUUUUUUUCAUCUUGCCCCUGGCCAGACAAAUGCCACGUCGGUUGACGACGGCACACUCGCCCA--AUGAAAUCUGCUCCUUGCCACGGGCG ..............................((((.((((((((..........((((.....))))(((......)))..--......))))......)))).)))). ( -27.00) >DroSec_CAF1 43653 96 + 1 UCUAUCUUUUUU-------UUUUUCAUCUUGCCCCUGGCCAGACAAAUGCCACGUCGGUUGA---AGGCACACUCGCCCA--AUGAAAUCUGCUCCUUGCCAGGGGCG ............-------...........(((((((((((((.....(((.....)))...---.(((......)))..--......))))......))))))))). ( -29.10) >DroSim_CAF1 46125 99 + 1 UCUAUUUUUUUUUCU----UUUUUCAUCUUGCCCCUGGCCAGACAAAUGCCACGUCGGUGGA---AGGCACACUCGCCCA--AUGAAAUCUGCUCCUUGCCAGGGGCG ...............----...........(((((((((((((......((((....)))).---.(((......)))..--......))))......))))))))). ( -32.50) >DroEre_CAF1 44663 100 + 1 UCCCAGUCUCUGU---UUUUUUUUCAUCUUGCCCCUGGCCAGACAAAUGCCACAUCGGUUGA---CGGCACACUCGUCCA--AGGAGAUCUACUCCUGGCCAGGGGCG .............---..............((((((((((.(((...((((.((.....)).---.)))).....)))..--(((((.....))))))))))))))). ( -38.00) >DroYak_CAF1 44461 101 + 1 UCUACUUUUUUUC---UUUUCAUUCAUCUUAG-CCUGGCCAGACAAAUGCCACGUCGCUUGA---CGGCACACUUGGCCAGAAGGAGAUCUACUCCUGGCCAGGGGCG .............---...............(-((((((.........))))((((....))---)).....(((((((((.((........)).)))))))))))). ( -31.00) >DroAna_CAF1 45790 81 + 1 AA---UUUUUU--------------GCCUUUCCC----ACAGACAAAUGCCACGUCACUUGA---CGGCACUC-GGCCCA--AUGCGGAUUCAUCCUAGCCAGGGGCG ..---.....(--------------(((......----.((......))....(((....))---)))))...-.((((.--..(((((....)))..))...)))). ( -19.20) >consensus UCUAUUUUUUUUU______UUUUUCAUCUUGCCCCUGGCCAGACAAAUGCCACGUCGGUUGA___CGGCACACUCGCCCA__AUGAAAUCUGCUCCUUGCCAGGGGCG ..............................(((((((((.((.....((((...((....))....))))..............((.......)))).))))))))). (-17.47 = -19.30 + 1.83)

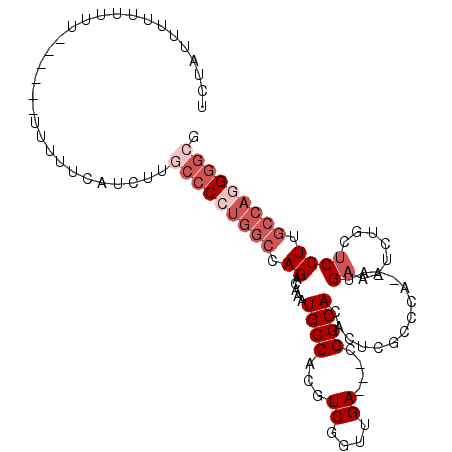

| Location | 10,048,014 – 10,048,120 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 75.81 |
| Mean single sequence MFE | -36.55 |
| Consensus MFE | -24.68 |
| Energy contribution | -26.85 |
| Covariance contribution | 2.17 |
| Combinations/Pair | 1.15 |
| Mean z-score | -3.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.47 |
| SVM RNA-class probability | 0.994382 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10048014 106 - 22407834 CGCCCGUGGCAAGGAGCAGAUUUCAU--UGGGCGAGUGUGCCGUCGUCAACCGACGUGGCAUUUGUCUGGCCAGGGGCAAGAUGAAAAAAAAUAGAAAAAAAAAUAGA .((((.((((..((((....))))..--.((((((((((..(((((.....)))))..)))))))))).)))).)))).............................. ( -38.50) >DroSec_CAF1 43653 96 - 1 CGCCCCUGGCAAGGAGCAGAUUUCAU--UGGGCGAGUGUGCCU---UCAACCGACGUGGCAUUUGUCUGGCCAGGGGCAAGAUGAAAAA-------AAAAAAGAUAGA .(((((((((..((((....))))..--.((((((((((..(.---((....)).)..)))))))))).)))))))))...........-------............ ( -35.30) >DroSim_CAF1 46125 99 - 1 CGCCCCUGGCAAGGAGCAGAUUUCAU--UGGGCGAGUGUGCCU---UCCACCGACGUGGCAUUUGUCUGGCCAGGGGCAAGAUGAAAAA----AGAAAAAAAAAUAGA .(((((((((..(((.(((((.....--.(((((....)))))---.((((....)))).)))))))).)))))))))...........----............... ( -35.10) >DroEre_CAF1 44663 100 - 1 CGCCCCUGGCCAGGAGUAGAUCUCCU--UGGACGAGUGUGCCG---UCAACCGAUGUGGCAUUUGUCUGGCCAGGGGCAAGAUGAAAAAAAA---ACAGAGACUGGGA .(((((((((((((((.....)))))--.((((((((((..((---((....))))..))))))))))))))))))))..............---.(((...)))... ( -49.30) >DroYak_CAF1 44461 101 - 1 CGCCCCUGGCCAGGAGUAGAUCUCCUUCUGGCCAAGUGUGCCG---UCAAGCGACGUGGCAUUUGUCUGGCCAGG-CUAAGAUGAAUGAAAA---GAAAAAAAGUAGA ....((((((((((((.....)))))...((.(((((((..((---((....))))..))))))).)))))))))-................---............. ( -34.80) >DroAna_CAF1 45790 81 - 1 CGCCCCUGGCUAGGAUGAAUCCGCAU--UGGGCC-GAGUGCCG---UCAAGUGACGUGGCAUUUGUCUGU----GGGAAAGGC--------------AAAAAA---UU .(((.((.((..(((..(((((((..--..(((.-....)))(---((....))))))).)))..)))))----.))...)))--------------......---.. ( -26.30) >consensus CGCCCCUGGCAAGGAGCAGAUCUCAU__UGGGCGAGUGUGCCG___UCAACCGACGUGGCAUUUGUCUGGCCAGGGGCAAGAUGAAAAA______AAAAAAAAAUAGA .(((((((((...(((.....))).....((((((..((((((...((....))..)))))))))))).))))))))).............................. (-24.68 = -26.85 + 2.17)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:49 2006