
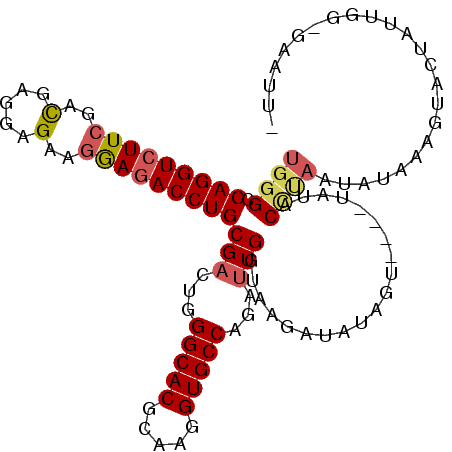
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,038,553 – 10,038,657 |
| Length | 104 |
| Max. P | 0.917586 |

| Location | 10,038,553 – 10,038,657 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -22.53 |
| Energy contribution | -23.03 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917586 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10038553 104 + 22407834 UGGGCCAGGUCUUCGACGAGGAGAAGGAGACCUGCGACUGGGCACGCAAGGUGCCAGAUUGGUAAGCUAUAGU----UAUCCCUAAUAUAAAGUACUAUUGG-GAAUUU ..((((((((((((..(.....)..)))))))))(((((((.(((.....))))))).)))....))).....----..((((.((((........))))))-)).... ( -32.90) >DroVir_CAF1 37790 94 + 1 UCGGCCAGGUGUUCGAUGAGGAGAACAAAACCUGCGACUGGGCACGUAAGGUGCCAGAUUGGUAAGAUUUGCCGUGUCGAACAAAAUGU-----------GGCGA---- ...((((.(((((((((..((..((....(((.....((((.(((.....)))))))...)))....))..))..))))))))...).)-----------)))..---- ( -30.30) >DroSec_CAF1 34429 104 + 1 UGGGCCAGGUCUUCGACGAGGAGAAGGAGACCUGCGACUGGGCACGCAAGGUGCCAGAUUGGUAAGCUAUAGU----UACACCUAAUAUAAAGUACUAUUCCUGAAUU- ((((.(((((((((..(.....)..))))))))).((((((((((.....)))))....(((....)))))))----)...)))).......................- ( -29.30) >DroSim_CAF1 36789 104 + 1 UGGGCCAGGUCUUCGACGAGGAGAAGGAGACCUGCGACUGGGCACGCAAGGUGCCAGAUUGGUAAGCUAUAGU----CACACCUAAUAUAAAGUAUUAUUGCUGAAUU- ((((.(((((((((..(.....)..))))))))).((((((((((.....)))))....(((....)))))))----)...))))......((((....)))).....- ( -33.50) >DroEre_CAF1 34388 104 + 1 UGGGCCAGGUCUUCGACGAGGAGAAGGAGACCUGCGACUGGGCACGCAAGGUGCCCGAUUGGUAAGAUAUGGU----UAUCCCCAAUAUUUAGUACUACCAG-AAGUUC (((((.((((((((..(.....)..)))))))))).(((((((((.....)))))).(((((...((((....----)))).)))))....)))....))).-...... ( -36.30) >DroYak_CAF1 35087 87 + 1 UGGGCCAGGUCUUCGACGAGGAGAAGGAGACCUGCGACUGGGCACGCAAGGUGCCCGACUGGUAAUAUAUGGU----UAUCCUUCCUACUU------------------ ((((((((((((((..(.....)..))))))))).)..(((((((.....)))))))...((((((.....))----))))...))))...------------------ ( -30.90) >consensus UGGGCCAGGUCUUCGACGAGGAGAAGGAGACCUGCGACUGGGCACGCAAGGUGCCAGAUUGGUAAGAUAUAGU____UAUACCUAAUAUAAAGUACUAUUGG_GAAUU_ ((((.(((((((((..(.....)..)))))))))(((...(((((.....)))))...)))....................))))........................ (-22.53 = -23.03 + 0.50)

| Location | 10,038,553 – 10,038,657 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.87 |
| Mean single sequence MFE | -25.33 |
| Consensus MFE | -16.47 |
| Energy contribution | -16.13 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.14 |
| Structure conservation index | 0.65 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10038553 104 - 22407834 AAAUUC-CCAAUAGUACUUUAUAUUAGGGAUA----ACUAUAGCUUACCAAUCUGGCACCUUGCGUGCCCAGUCGCAGGUCUCCUUCUCCUCGUCGAAGACCUGGCCCA ....((-((((((........)))).))))..----......((........(((((((.....))).))))...(((((((.(..(.....)..).)))))))))... ( -24.80) >DroVir_CAF1 37790 94 - 1 ----UCGCC-----------ACAUUUUGUUCGACACGGCAAAUCUUACCAAUCUGGCACCUUACGUGCCCAGUCGCAGGUUUUGUUCUCCUCAUCGAACACCUGGCCGA ----..(((-----------(.....(((((((...((..(((...(((...(((((((.....))).)))).....)))...)))..))...)))))))..))))... ( -25.50) >DroSec_CAF1 34429 104 - 1 -AAUUCAGGAAUAGUACUUUAUAUUAGGUGUA----ACUAUAGCUUACCAAUCUGGCACCUUGCGUGCCCAGUCGCAGGUCUCCUUCUCCUCGUCGAAGACCUGGCCCA -......((.(((((((((......))))...----))))).((........(((((((.....))).))))...(((((((.(..(.....)..).))))))))))). ( -23.70) >DroSim_CAF1 36789 104 - 1 -AAUUCAGCAAUAAUACUUUAUAUUAGGUGUG----ACUAUAGCUUACCAAUCUGGCACCUUGCGUGCCCAGUCGCAGGUCUCCUUCUCCUCGUCGAAGACCUGGCCCA -......((....((((((......))))))(----(((...(.....).....(((((.....))))).)))).(((((((.(..(.....)..).)))))))))... ( -22.70) >DroEre_CAF1 34388 104 - 1 GAACUU-CUGGUAGUACUAAAUAUUGGGGAUA----ACCAUAUCUUACCAAUCGGGCACCUUGCGUGCCCAGUCGCAGGUCUCCUUCUCCUCGUCGAAGACCUGGCCCA ......-..(((.(.(((....((((((((((----....)))))..))))).((((((.....)))))))))).(((((((.(..(.....)..).)))))))))).. ( -30.40) >DroYak_CAF1 35087 87 - 1 ------------------AAGUAGGAAGGAUA----ACCAUAUAUUACCAGUCGGGCACCUUGCGUGCCCAGUCGCAGGUCUCCUUCUCCUCGUCGAAGACCUGGCCCA ------------------.....((..((...----.))........((((((((((((.....)))))).(.((.(((.........))))).)...)).)))).)). ( -24.90) >consensus _AAUUC_CCAAUAGUACUUUAUAUUAGGGAUA____ACCAUAGCUUACCAAUCUGGCACCUUGCGUGCCCAGUCGCAGGUCUCCUUCUCCUCGUCGAAGACCUGGCCCA ..........................((..........................(((((.....)))))......(((((...((((........)))))))))..)). (-16.47 = -16.13 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:44 2006