
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,034,635 – 10,034,827 |
| Length | 192 |
| Max. P | 0.709775 |

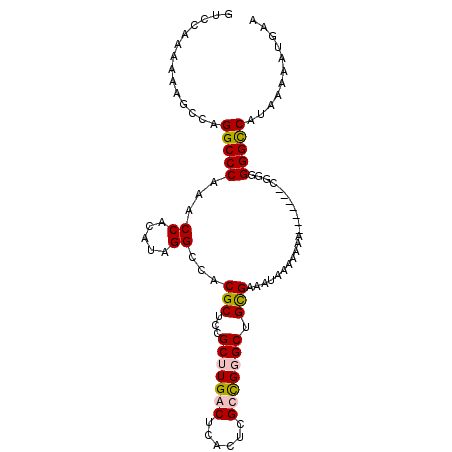
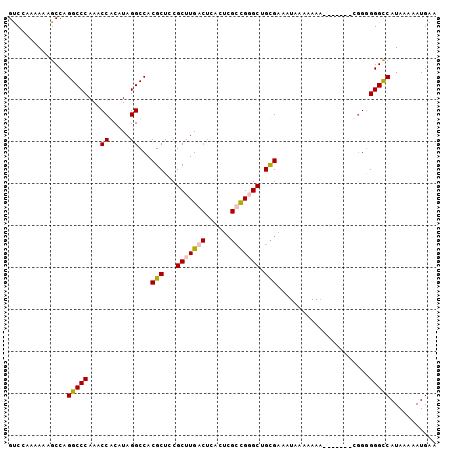
| Location | 10,034,635 – 10,034,737 |
|---|---|
| Length | 102 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 82.06 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -20.09 |
| Energy contribution | -20.52 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.709775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10034635 102 + 22407834 GUCGAAAAAAGCCAGGCCCAAACCGCAUAGGCCACGCUUCGCUUGACUCGUUCGCCGGGCUGCGAAAUAAAAAAAAAAAAAACGGAGGGUCAUAAAAAUGAA ..............(((((...((((....)).....(((((..(.((((.....))))).))))).................)).)))))........... ( -20.10) >DroSim_CAF1 32933 86 + 1 GUCGAAAAAUGCCAGGCCCAAACCACAUAGGCCACGCUCCGCUUGA---------CGGGCUGUGAAAUAAAAAAA-------AGGGGGGCCAUAAAAAUGAA ..............(((((...((.....)).(((((.(((.....---------))))).)))...........-------....)))))........... ( -20.80) >DroEre_CAF1 30419 93 + 1 GUCCAAAAAAGCCAGGCCCAAACCACAUAGGCCACGCUCCGCUUGGCUCACUCGCUGUGCUGCGAAAUAAAAAA--------C-GGGGGCCAUAAAAAUGAA ..............(((((...((.....(((((.((...)).)))))...((((......)))).........--------.-)))))))........... ( -25.30) >DroYak_CAF1 31021 95 + 1 GCCCAAAAAAGCCAGGCCCAAACCGCAUAGGCCACGCUUCGCUUGGCUCACUCGCUGUGCGGCGAUAUAAAAAAA-------CGGGGGGCCAUAAAAAUGAA ..............(((((...(((....(((((.((...)).)))))...(((((....)))))..........-------))).)))))........... ( -30.20) >consensus GUCCAAAAAAGCCAGGCCCAAACCACAUAGGCCACGCUCCGCUUGACUCACUCGCCGGGCUGCGAAAUAAAAAAA_______CGGGGGGCCAUAAAAAUGAA ..............(((((...((.....))...(((...(((((((......))))))).)))......................)))))........... (-20.09 = -20.52 + 0.44)

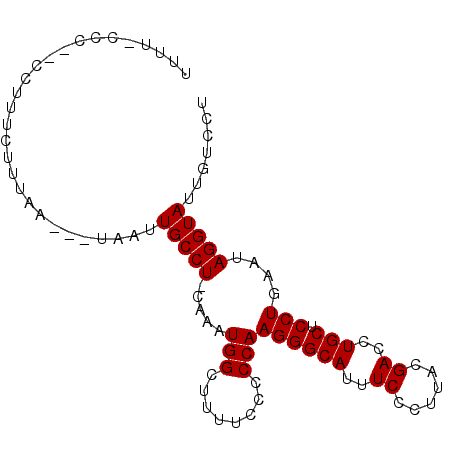
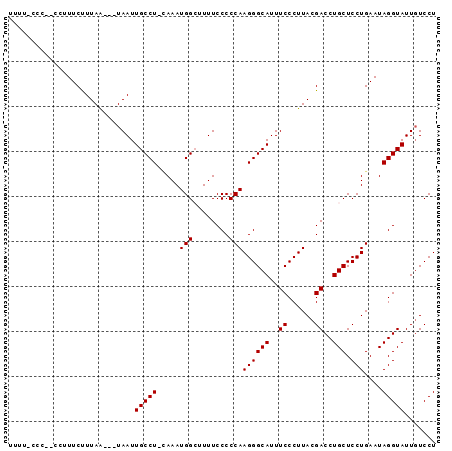
| Location | 10,034,737 – 10,034,827 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 90.74 |
| Mean single sequence MFE | -14.62 |
| Consensus MFE | -11.86 |
| Energy contribution | -11.86 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.628142 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10034737 90 - 22407834 UUUU-CCCAGCCAUUCUUUAA---UAAUUGCCU-GAAAUGGCUUUUCCCCCAAGGGCAUUUCCCUUACGACCUGCUCCUGAAUAGGUAUUUUCCU ....-....(((((((.....---.....(((.-.....)))..........((((((..((......))..))).))))))).)))........ ( -16.70) >DroSec_CAF1 30657 88 - 1 UUUU-CCC--CCUUUCCUUAA---UAAUUGCCU-CAAAUGGCUUUUCCCCCAAGGGCAUUUCCCUUACGACCUGCUCCUGAAUAGGUAUUGUCCU ....-...--((((((.....---.....(((.-.....)))..........((((((..((......))..))).)))))).)))......... ( -14.00) >DroSim_CAF1 33019 88 - 1 UUUC-CCC--CCUUUCUUUAA---UAAUUGCCU-CAAAUGGCUUUUCCCCCAAGGGCAUUUCCCUUACGACCUGCUCCUGAAUAGGUAUUGUCCU ....-...--((((((.....---.....(((.-.....)))..........((((((..((......))..))).)))))).)))......... ( -14.40) >DroEre_CAF1 30512 91 - 1 UUUU-UCU--CAUUUCCUUAAUAAUAAUUGCCUGUAAAUGG-UUUUCCCCCAAGGGCAUUUCCCUUACGACCUGCUCCUGAAUAGGUAUUGUCCU ....-...--.............(((((.((((((...(((-.......)))((((((..((......))..))).)))..)))))))))))... ( -15.40) >DroYak_CAF1 31116 88 - 1 UGUUUCCU--CGUUUCUUUAG---UAAUUGCCU-CAAAUGG-UUUUCCCCCAAGGGCAUUUCCCUUACGACCUGCUCCUGAAUAGGUAUUGUCCU .....(((--...(((...((---((...(((.-.....))-)........(((((.....)))))......))))...))).)))......... ( -12.60) >consensus UUUU_CCC__CCUUUCUUUAA___UAAUUGCCU_CAAAUGGCUUUUCCCCCAAGGGCAUUUCCCUUACGACCUGCUCCUGAAUAGGUAUUGUCCU ............................(((((.....(((........)))((((((..((......))..))).)))....)))))....... (-11.86 = -11.86 + 0.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:42 2006