
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 10,030,555 – 10,030,671 |
| Length | 116 |
| Max. P | 0.561060 |

| Location | 10,030,555 – 10,030,671 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 81.95 |
| Mean single sequence MFE | -28.19 |
| Consensus MFE | -19.89 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.561060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 10030555 116 + 22407834 GCAGUGCCAUUUCCAUGGCCAUUAUAUUCAAUUAUACGACAAUAAUGUGGCGCUUAGGGCGCUGCGGUUCACGGGAAUUGAAAAUAAUAAUAAUAUGGGGCGGCGGGGGGGUGUUG .(((..((.(..((.(.((((((((.(((((((...((.......((..((((.....))))..)).....))..))))))).)))))..........))).).))..)))..))) ( -36.50) >DroPse_CAF1 24622 94 + 1 ------------CCAUUGCCAUUAUAUUCAAUUAUACGACAAUAACGUGGCGCUUAGGGCGCUGCGGUUACCAGAAAUUGAAAAUAAUAAUAAAGCGG--------AGGAGGAU-- ------------((.((((.(((((.(((((((............((..((((.....))))..)).........))))))).)))))......))))--------.)).....-- ( -22.00) >DroSim_CAF1 27839 108 + 1 GCAGUGCCAUUUCCAUGGCCAUUAUAUUCAAUUAUACGACAAUAACGUGGCGCUUAGGGCGCUGCGGUUCACGGGAAUUGAAAAUAAUAAUAAAAUGG--------GGGGUUGUUU (((..(((...(((((....(((((.(((((((...((.......((..((((.....))))..)).....))..))))))).)))))......))))--------).)))))).. ( -30.30) >DroEre_CAF1 26240 107 + 1 GCAGUGCCAUUUCCAUGGCCAUUAUAUUCAAUUAUACGACAAUAACGUGGCGCUUAGGGCGCUGCGGUUCACGGGAAUUGAAAAUAAUAAUAAUGUGG--------GGGGU-GUAG ...(..((...((((((...(((((.(((((((...((.......((..((((.....))))..)).....))..))))))).))))).....)))))--------).)).-.).. ( -31.60) >DroAna_CAF1 26857 107 + 1 GCAGCGUCAUUUCCAUGGCCAUUAUAUUCAAUUAUACGACAAUAACGUGGCGCUUAGGGCGCUGCGGUUCACAGGAAUUGAAAAUAAUAAUAAAGCGA--------AGGGA-CCAG ((.(.(((((....))))))(((((.(((((((...(........((..((((.....))))..)).......).))))))).)))))......))..--------.....-.... ( -26.76) >DroPer_CAF1 25513 94 + 1 ------------CCAUUGCCAUUAUAUUCAAUUAUACGACAAUAACGUGGCGCUUAGGGCGCUGCGGUUACCAGAAAUUGAAAAUAAUAAUAAAGCGG--------AGGAGGAU-- ------------((.((((.(((((.(((((((............((..((((.....))))..)).........))))))).)))))......))))--------.)).....-- ( -22.00) >consensus GCAGUGCCAUUUCCAUGGCCAUUAUAUUCAAUUAUACGACAAUAACGUGGCGCUUAGGGCGCUGCGGUUCACAGGAAUUGAAAAUAAUAAUAAAGCGG________AGGGG_GU_G ....................(((((.(((((((............((..((((.....))))..)).........))))))).)))))............................ (-19.89 = -19.75 + -0.14)
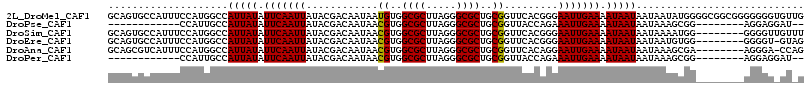
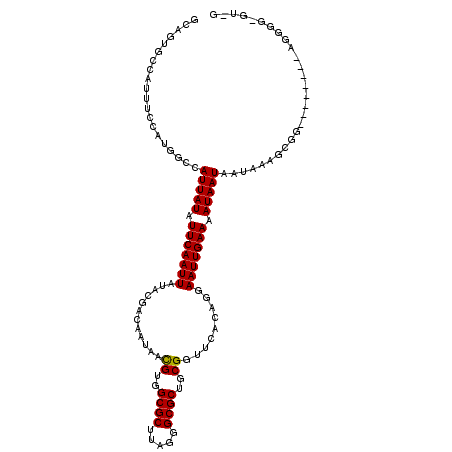
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:40 2006