
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,994,595 – 9,994,703 |
| Length | 108 |
| Max. P | 0.576276 |

| Location | 9,994,595 – 9,994,703 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -42.18 |
| Consensus MFE | -33.12 |
| Energy contribution | -33.60 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.65 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.576276 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
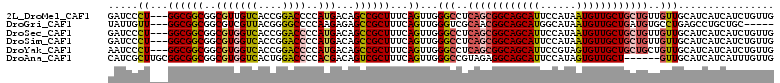
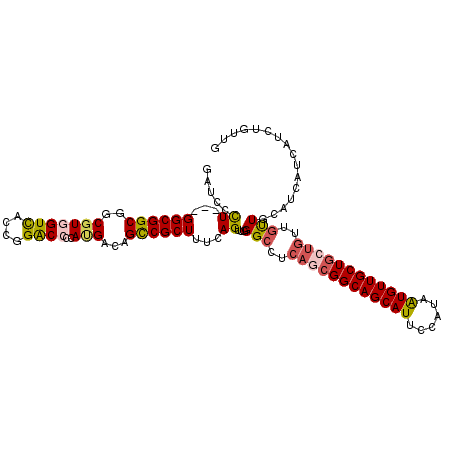
>2L_DroMel_CAF1 9994595 108 - 22407834 GAUCCCU---GGCGGCGGCGUUGUCACCGGACCCCAUGACAGCCGCUUUCAGUUGGGCCUCAGCGGCAGCAUUCCAUAAUGUUGCUGCUGUUGUUGCAUCAUCAUCUGUUG (((((((---((.((((((..(((((..(....)..))))))))))).))))..))((..(((((((((((...........)))))))))))..)))))........... ( -43.00) >DroGri_CAF1 29395 103 - 1 UAUUGUU---GGCGGCGGCGUCGUUACGGGGCCCCAAGAGAGCCGCUUUCAGUUGGGUCGCAACGGCAGCAUGGCAUAAUGUUGCUGAUGUGCCUGAGCCUGCUGC----- ....((.---(((((((((.((.....((....))....)))))))).....(..((.((((.(((((((((......))))))))).))))))..)))).))...----- ( -43.70) >DroSec_CAF1 25910 108 - 1 GAUCCCU---GGCGGCGGCGUGGUCACCGGACCCCAUGACAGCCGCUUUCAGUUGGGCCUCAGCGGCAGCAUUCCAUAAUGUUGCUGCUGUUGUUGCAUCAUCAUCUGUUG (((((((---((.((((((...((((..(....)..)))).)))))).))))..))((..(((((((((((...........)))))))))))..)))))........... ( -43.70) >DroSim_CAF1 28790 108 - 1 GAUCCCU---GGCGGCGGCGUGGUCACCGGACCCCAUGACAGCCGCUUUCAGUUGGGCCUCAGCGGCAGCAUUCCAUAAUGUUGCUGCUGUUGUUGCAUCAUCAUCUGUUG (((((((---((.((((((...((((..(....)..)))).)))))).))))..))((..(((((((((((...........)))))))))))..)))))........... ( -43.70) >DroYak_CAF1 26666 108 - 1 AAUCCCU---GGCGGCGGCGUGGUCACCGGACCCCAUGACAGCCGCUUUCAGUUGGGCCUCAGCGGCAGCAUUCCGUAGUGUUGCUGCUGCUGUUGCAUCAUCAUCUGUUG ...((((---((.((((((...((((..(....)..)))).)))))).))))..))((..(((((((((((...........)))))))))))..)).............. ( -45.60) >DroAna_CAF1 25974 105 - 1 CAUCGCUUGCGGCGGCGGCGUGGUCACUGGACCCCACGACAGUCGCUUUCAGUUGGGCCGUAGAGGCAGCAUUCCAUAGUGUUGCU------GUUGCAUCAUCAUUUGUUG ....(((((((((((((((...(((..(((...))).))).)))))).((....))))))))).((((((((......))))))))------...)).............. ( -33.40) >consensus GAUCCCU___GGCGGCGGCGUGGUCACCGGACCCCAUGACAGCCGCUUUCAGUUGGGCCUCAGCGGCAGCAUUCCAUAAUGUUGCUGCUGUUGUUGCAUCAUCAUCUGUUG .....((...((((((..(((((((....))))..)))...))))))...))...(((..((((((((((((......))))))))))))..)))................ (-33.12 = -33.60 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:33 2006