
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
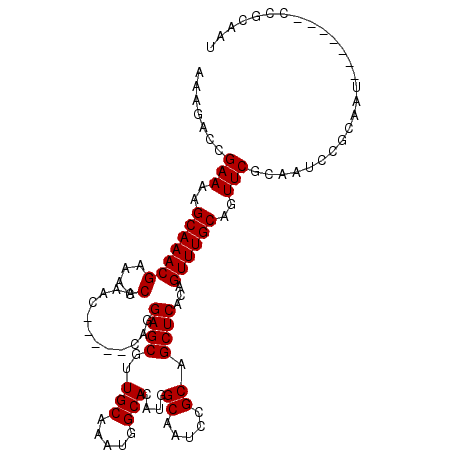
| Location | 9,991,040 – 9,991,131 |
| Length | 91 |
| Max. P | 0.884193 |

| Location | 9,991,040 – 9,991,131 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -18.30 |
| Energy contribution | -18.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.584469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9991040 91 + 22407834 -AAGACCGAAAAGCAAACGCAACCAAAC-----CAGGAGCAUUGCAAAUGGCACAUGGCAAUCCGCAGCUCACAGUUUGCAGUUCGCAAUC--------------CGCAAU -......(((..((((((.....(....-----..)((((..(((.....((.....)).....)))))))...))))))..)))((....--------------.))... ( -19.90) >DroSec_CAF1 22316 99 + 1 AAAGACCGAAAAGCAAACGAAAGCAAAC-----CAGGAGCGUUGCAAAUGGCACAUGGCAAUCCGCAGCUCACAGUUUGCAGUUCGCAAUCCGCAAU-------CCGCAAU ............((....(((.((((((-----...((((..(((.....((.....)).....)))))))...))))))..)))((.....))...-------..))... ( -23.40) >DroSim_CAF1 25190 99 + 1 AAAGACCGAAAAGCAAACGAAAGCAAAC-----CAGGAGCGUUGCAAAUGGCACAUGGCAAUCCGCAGCUCACAGUUUGCAGUUCGCAAUCCGCAAU-------CCGCAAU ............((....(((.((((((-----...((((..(((.....((.....)).....)))))))...))))))..)))((.....))...-------..))... ( -23.40) >DroYak_CAF1 23092 111 + 1 AAAGACAGAAAAGCAAACGAAACCAAACCAAACCAAGAGCGUUGCAAAUGGCACAUGGCUAUCCGCAGCUCACAGUUUGCAGUUCGCAAUCUGAAAUCCGCAAUCCACAAU ............((......................((((..(((..(((((.....)))))..))))))).(((.((((.....)))).)))......)).......... ( -23.30) >consensus AAAGACCGAAAAGCAAACGAAACCAAAC_____CAGGAGCGUUGCAAAUGGCACAUGGCAAUCCGCAGCUCACAGUUUGCAGUUCGCAAUCCGCAAU_______CCGCAAU .......(((..(((((((....)............((((..(((.....)))....((.....)).))))...))))))..))).......................... (-18.30 = -18.30 + 0.00)

| Location | 9,991,040 – 9,991,131 |
|---|---|
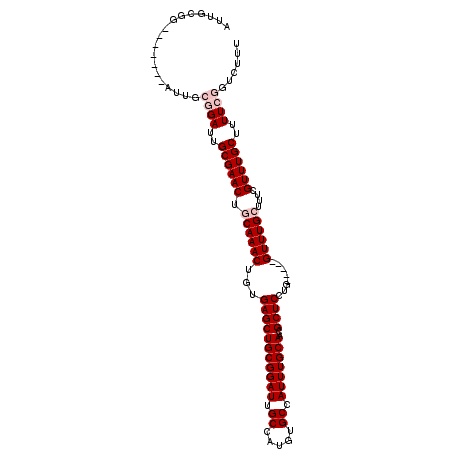
| Length | 91 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 86.35 |
| Mean single sequence MFE | -33.38 |
| Consensus MFE | -23.08 |
| Energy contribution | -24.57 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9991040 91 - 22407834 AUUGCG--------------GAUUGCGAACUGCAAACUGUGAGCUGCGGAUUGCCAUGUGCCAUUUGCAAUGCUCCUG-----GUUUGGUUGCGUUUGCUUUUCGGUCUU- ....((--------------((..((((((.((((((((.(((((((((((.((.....)).)))))))..)))).))-----))....))))))))))..)))).....- ( -31.00) >DroSec_CAF1 22316 99 - 1 AUUGCGG-------AUUGCGGAUUGCGAACUGCAAACUGUGAGCUGCGGAUUGCCAUGUGCCAUUUGCAACGCUCCUG-----GUUUGCUUUCGUUUGCUUUUCGGUCUUU .....((-------((..((((..((((((.((((((((.(((((((((((.((.....)).)))))))..)))).))-----))))))....))))))..)))))))).. ( -36.30) >DroSim_CAF1 25190 99 - 1 AUUGCGG-------AUUGCGGAUUGCGAACUGCAAACUGUGAGCUGCGGAUUGCCAUGUGCCAUUUGCAACGCUCCUG-----GUUUGCUUUCGUUUGCUUUUCGGUCUUU .....((-------((..((((..((((((.((((((((.(((((((((((.((.....)).)))))))..)))).))-----))))))....))))))..)))))))).. ( -36.30) >DroYak_CAF1 23092 111 - 1 AUUGUGGAUUGCGGAUUUCAGAUUGCGAACUGCAAACUGUGAGCUGCGGAUAGCCAUGUGCCAUUUGCAACGCUCUUGGUUUGGUUUGGUUUCGUUUGCUUUUCUGUCUUU .....(((..((((((....((...(((((..(((((((.(((((((((((.((.....)).)))))))..)))).))))))))))))...))))))))..)))....... ( -29.90) >consensus AUUGCGG_______AUUGCGGAUUGCGAACUGCAAACUGUGAGCUGCGGAUUGCCAUGUGCCAUUUGCAACGCUCCUG_____GUUUGCUUUCGUUUGCUUUUCGGUCUUU ..................((((..((((((.((((((...(((((((((((.((.....)).)))))))..))))........))))))....))))))..))))...... (-23.08 = -24.57 + 1.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:31 2006