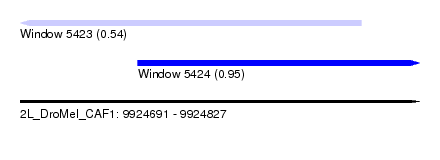
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,924,691 – 9,924,827 |
| Length | 136 |
| Max. P | 0.953471 |

| Location | 9,924,691 – 9,924,807 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.66 |
| Mean single sequence MFE | -41.87 |
| Consensus MFE | -26.51 |
| Energy contribution | -25.22 |
| Covariance contribution | -1.30 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.542877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
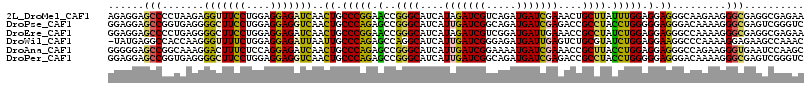
>2L_DroMel_CAF1 9924691 116 - 22407834 AGAGGAGCCCCUAAGAGGUUUCCUGGAGGAGAUCAACUGCCCGGAACCGGGCAUCAUAGAUCGUCAGAUGAUCGAAACUGCUUAUUUGGAGGAGGGCAAGAAGGGCGAGGCGAGAA ......(((((((.(((((((((....)))))))...((((((....)))))))).))).((..(((((((.((....)).)))))))..)).))))................... ( -36.90) >DroPse_CAF1 1791 116 - 1 GGAGGAGCCGGUGAGGGGCUUCCUGGAGGAGGUCAACUGCCCAGAGCCGGGCAUCAUUGAUCGGCAGAUGAUCGAGACCGCCUACCUGGGGGAGGGACAAAAGGGCGAGUCGGGUC ......((((((.((.(((((((....)))))))...(((((......)))))...)).))))))....((((..((((((((.(((.....))).......))))).))).)))) ( -44.90) >DroEre_CAF1 1890 116 - 1 GGAGGAGCCCCUGAGGGGCUUCCUGGAGGAGAUCAACUGCCCGGAACCGGGCAUCAUAGAUCGUCGGAUGAUUGAAACCGCCUAUCUGGAGGAGGGCCAAAAGGGCGAGGCGAGAA ((.((((((((...))))))))((((....((((...((((((....)))))).....)))).))))..........))((((.(((.....)))(((.....))).))))..... ( -44.10) >DroWil_CAF1 2116 115 - 1 -UAUGAGGCCACCAAGGGUUUUCUGGAGGAGAUUAAUUGCCCAGAGCCAGGCAUCAUUGAUCGGGAGAUGAUUGAGUCUGCGUAUCUGGAGGAAGGCCCAAAAGGAGAAGCCAAAC -.....(((..((..((((((((((.((........)).)((((((((((((.(((.(.(((....))).).)))))))).)).))))).)))))))))....))....))).... ( -37.00) >DroAna_CAF1 3128 116 - 1 GGGGGAGCCGGCAAAGGACUUUCUCCAGGAGAUCAACUGCCCAGAGCCGGGCAUCAUUGAUCGGAAAAUGAUCGAAACCGCUUACCUGGAGGAGGGCCAGAAGGGUGAAUCCAAGC (((...(((......((.(((((((((((.((((((.(((((......)))))...))))))......(((.((....)).)))))))))))))).)).....)))...))).... ( -43.40) >DroPer_CAF1 1791 116 - 1 GGAGGAGCCGGUGAGGGGCUUCCUGGAGGAGGUCAACUGCCCAGAGCCGGGCAUCAUUGAUCGGCAGAUGAUCGAGACCGCCUACCUGGGGGAGGGACAAAAGGGCGAGUCGGGUC ......((((((.((.(((((((....)))))))...(((((......)))))...)).))))))....((((..((((((((.(((.....))).......))))).))).)))) ( -44.90) >consensus GGAGGAGCCCGUGAGGGGCUUCCUGGAGGAGAUCAACUGCCCAGAGCCGGGCAUCAUUGAUCGGCAGAUGAUCGAAACCGCCUACCUGGAGGAGGGCCAAAAGGGCGAGGCGAGAC ......(((.......(((((((....)))))))..((.(((((.(..((((....(((((((.....)))))))....)))).))))).).)).........))).......... (-26.51 = -25.22 + -1.30)

| Location | 9,924,731 – 9,924,827 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 75.53 |
| Mean single sequence MFE | -26.48 |
| Consensus MFE | -18.34 |
| Energy contribution | -18.95 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953471 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
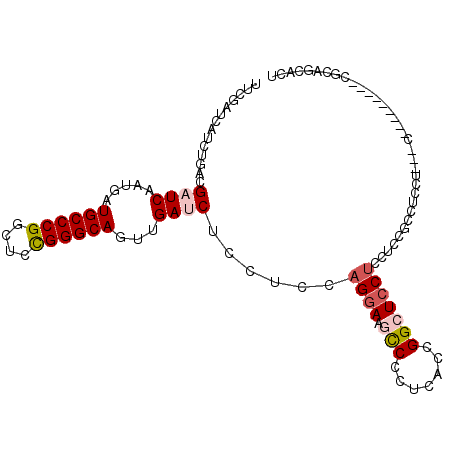
>2L_DroMel_CAF1 9924731 96 + 22407834 UUCGAUCAUCUGACGAUCUAUGAUGCCCGGUUCCGGGCAGUUGAUCUCCUCCAGGAAACCUCUUAGGGGCUCCUCUUCCGUCUCCU---C---------CACAGCAUC ...........(((((((.....((((((....))))))...))))......((((..(((....)))..)))).....)))....---.---------......... ( -27.90) >DroPse_CAF1 1831 92 + 1 CUCGAUCAUCUGCCGAUCAAUGAUGCCCGGCUCUGGGCAGUUGACCUCCUCCAGGAAGCCCCUCACCGGCUCCUCCAU---CUCAU---C---------CCCCGAAC- ...(((.....((((.(((((..((((((....)))))))))))..(((....)))..........))))......))---)....---.---------........- ( -22.30) >DroSec_CAF1 1934 105 + 1 UUCGAUCAUCUGACGAUCUAUGAUGCCCGGUUCCGGGCAGUUGAUCUCCUCCAGGAAACCCAUUAGGGGCUCCUCCUCCGCCUCCU---CCUCCUUUUCCGUAGCACU ............((((((.....((((((....))))))...)))).......(((((......((((((.........))).)))---......)))))))...... ( -29.10) >DroEre_CAF1 1930 102 + 1 UUCAAUCAUCCGACGAUCUAUGAUGCCCGGUUCCGGGCAGUUGAUCUCCUCCAGGAAGCCCCUCAGGGGCUCCUCCUCCGCCUCCU------UCUCCUCCGCAGCACU ...........((.((((.....((((((....))))))...))))))....((((.(((((...)))))))))............------................ ( -30.90) >DroAna_CAF1 3168 108 + 1 UUCGAUCAUUUUCCGAUCAAUGAUGCCCGGCUCUGGGCAGUUGAUCUCCUGGAGAAAGUCCUUUGCCGGCUCCCCCUCCUCCUCCAACUCCUCGACUCCCGCCUCACU .((((.........(((((((..((((((....)))))))))))))....((((..((((.......))))....))))............))))............. ( -26.40) >DroPer_CAF1 1831 92 + 1 CUCGAUCAUCUGCCGAUCAAUGAUGCCCGGCUCUGGGCAGUUGACCUCCUCCAGGAAGCCCCUCACCGGCUCCUCCAU---CUCAU---C---------CCCCGAAC- ...(((.....((((.(((((..((((((....)))))))))))..(((....)))..........))))......))---)....---.---------........- ( -22.30) >consensus UUCGAUCAUCUGACGAUCAAUGAUGCCCGGCUCCGGGCAGUUGAUCUCCUCCAGGAAGCCCCUCACCGGCUCCUCCUCCGCCUCCU___C_________CGCAGCACU ..............((((.....((((((....))))))...))))......((((.(((.......))))))).................................. (-18.34 = -18.95 + 0.61)

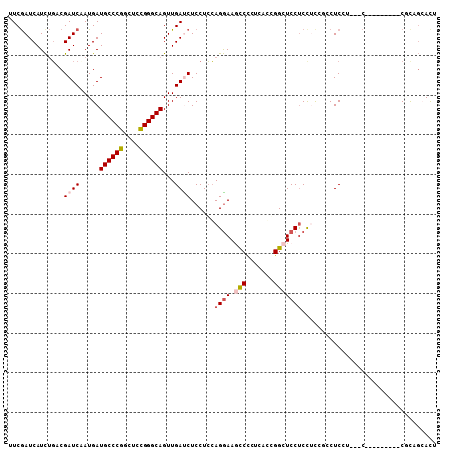
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:14 2006