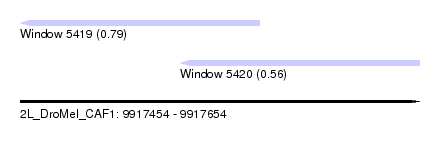
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,917,454 – 9,917,654 |
| Length | 200 |
| Max. P | 0.793407 |

| Location | 9,917,454 – 9,917,574 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.39 |
| Mean single sequence MFE | -32.85 |
| Consensus MFE | -19.67 |
| Energy contribution | -19.23 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.42 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.793407 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
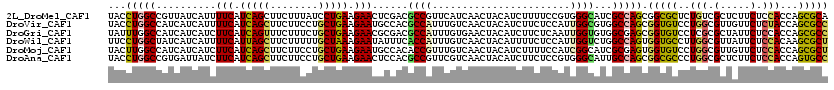
>2L_DroMel_CAF1 9917454 120 - 22407834 UACCUGGCCGUUAUCAUUUUCAUCAGCUUCUUUAUCCUGAAGAACUCGACGCCGUUCAUCAACUACAUCUUUUCCGUGGGCAUCGCCAGCGGCGCUCUGUCGCUCUUCUCCACCAGCGCA ......((((((......(((.((((..........)))).)))..(((.((((((....)))..(((.......)))))).)))..))))))((.(((..(........)..))).)). ( -23.80) >DroVir_CAF1 15291 120 - 1 UACCUGGCCAUCAUCAUUUUCAUCAGCUUCUUCCUGCUGAAGAAUGCCACGCCAUUUGUCAACUACAUCUUCUCCAUUGGCGUGGCCAGCGGUGUCCUGGCGUUGUUCUCUACCAGCGCC .....((.((((......(((.(((((........))))).))).(((((((((.......................)))))))))....)))).)).(((((((........))))))) ( -41.80) >DroGri_CAF1 14681 120 - 1 UAUUUGGCCAUCAUCAUCUUCAUCAGUUUCUUUCUGCUGAAGAACGCGACGCCAUUUGUGAACUACAUCUUCUCAAUUGGUGUGGCGAGCGGUGUCCUCGCGCUAUUCUCCACCAGCGCC .....((.((((....((((((.(((.......))).)))))).(((.((((((.(((.(((.......))).))).)))))).)))...)))).))..(((((..........))))). ( -41.50) >DroWil_CAF1 31727 120 - 1 UUCCUGGCUAUCAUCAUUUUCAUUAGCUUCUUUUUGCUAAAGAAUAUUUCACCAUUUGUCAACUACAUUUUCUCCAUUGGUCUGGCCAGUGGUGCCUUGGCGUUAUUCUCCACAAGCGCU .....(((..........(((.(((((........))))).)))......((((((.((((((((............)))).)))).)))))))))..((((((..........)))))) ( -26.80) >DroMoj_CAF1 16470 120 - 1 UACUUGGCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAAUGCCACACCGUUUGUCAACUACAUCUUUUCCAUCGGCAUCGCGAGUGGUGUCCUGGCGUUGUUCUCCACCAGCGCU .....((.(((((((.((((((.(((.......))).))))))(((((........(((.....)))...........)))))...)).))))).)).(((((((........))))))) ( -28.71) >DroAna_CAF1 13948 120 - 1 UACCUGGCCGUGAUUAUCUUCAUCAGCUUCUUCCUGCUGAAGAACUCCACGCCGUUCGUCAACUACAUCUUCUCCGUGGGCAUUGCCAGCGGCGCCCUGGCGCUCUUCUCCACCAGUGCC .....(((.(((....((((((.(((.......))).))))))....)))((((((.((...((((.........)))).....)).)))))))))..((((((..........)))))) ( -34.50) >consensus UACCUGGCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGAAGAACGCCACGCCAUUUGUCAACUACAUCUUCUCCAUUGGCAUGGCCAGCGGUGCCCUGGCGCUAUUCUCCACCAGCGCC ...(((((..........(((.(((((........))))).)))......((((.......................))))...))))).(((((..(((.(.....).)))...))))) (-19.67 = -19.23 + -0.44)
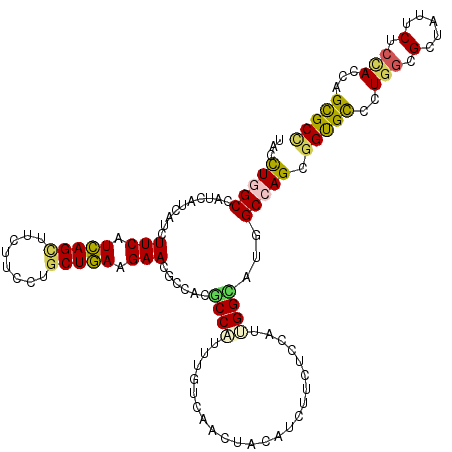
| Location | 9,917,534 – 9,917,654 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Mean single sequence MFE | -31.71 |
| Consensus MFE | -23.92 |
| Energy contribution | -23.45 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.558321 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9917534 120 - 22407834 ACGAGCGCAUCCUGUGGAAGAAGAACGAGGUAGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAACAACGCCAUCUACCUGGCCGUUAUCAUUUUCAUCAGCUUCUUUAUCCUGA ..((((......(((((((((((((.(.(((.(((........))).))).).))))...)))))))))..(((((((......))).)))).............))))........... ( -32.90) >DroVir_CAF1 15371 120 - 1 ACGAACGCAUCCUGUGGAAGAAGAACGAGGUUGCCGACUAUGAAGCGACCACAUACUCGAUUUUCUAUAACAACGCCCUCUACCUGGCCAUCAUCAUUUUCAUCAGCUUCUUCCUGCUGA ..((((((.....)))((((((((.(((((((((..........))))))......))).))))))........(((........)))..))......))).(((((........))))) ( -24.80) >DroGri_CAF1 14761 120 - 1 ACGAACGCAUCCUGUGGAAGAAGAACGAGGUGGCUGACUAUGAGGCAACCACAUAUUCGAUUUUCUACAACAAUGCCAUCUAUUUGGCCAUCAUCAUCUUCAUCAGUUUCUUUCUGCUGA .....(((.....)))...(((((..((((((((..(.((.((((((..........................)))).)))).)..)))))).)).))))).(((((........))))) ( -28.77) >DroWil_CAF1 31807 120 - 1 AUGAACGCAUCUUGUGGAAAAAGAACGAAGUGGCUGACUAUGAAGCCACCACGUAUUCGAUUUUCUAUAACAAUGCCCUCUUCCUGGCUAUCAUCAUUUUCAUUAGCUUCUUUUUGCUAA (((((.((((.((((((((((.(((((..((((((........))))))..)))..))..))))))))))..))))((.......))...........)))))((((........)))). ( -30.00) >DroMoj_CAF1 16550 120 - 1 ACGAACGCAUCCUGUGGAAGAAGAAUGAAGUUGCCGACUACGAGGCCACGACCUAUUCGAUUUUCUACAACAAUGCCAUCUACUUGGCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGA ..(((.......(((((((((.(((((..((.(((........))).))....)))))..))))))))).....((((......))))..........))).(((((........))))) ( -30.70) >DroAna_CAF1 14028 120 - 1 AUGAGCGUAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUUCUCCAUCUUCUACAAUAACGCCAUCUACCUGGCCGUGAUUAUCUUCAUCAGCUUCUUCCUGCUGA .((((.(((((((((((((((((((.(.(((((((........))))))).).))))...))))))))).....((((......)))).).)).))).))))(((((........))))) ( -43.10) >consensus ACGAACGCAUCCUGUGGAAGAAGAACGAGGUGGCCGACUACGAGGCCACCACCUACUCGAUUUUCUACAACAACGCCAUCUACCUGGCCAUCAUCAUCUUCAUCAGCUUCUUCCUGCUGA ..(((.......(((((((((.((..(.(((.(((........))).))).)....))..))))))))).....((((......))))..........))).(((((........))))) (-23.92 = -23.45 + -0.47)

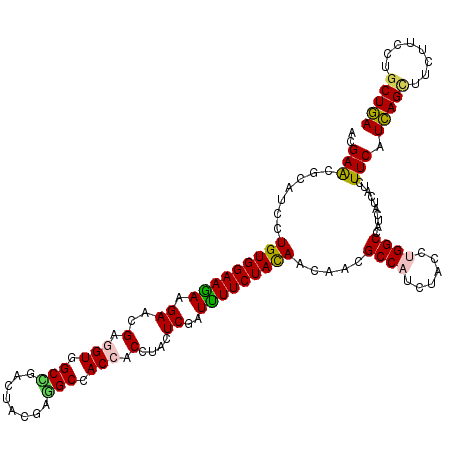
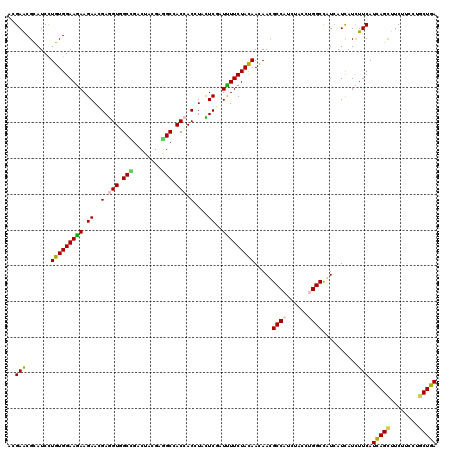
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:10 2006