
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,903,831 – 9,903,973 |
| Length | 142 |
| Max. P | 0.851002 |

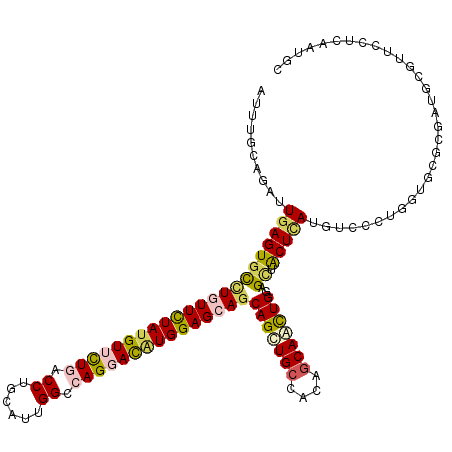
| Location | 9,903,831 – 9,903,949 |
|---|---|
| Length | 118 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -41.25 |
| Consensus MFE | -29.42 |
| Energy contribution | -29.57 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9903831 118 - 22407834 ACUUUCAGAUUGAGUGCUUGUUUUAUGUUUUGACCUGUAUUGGCCAGGACAUGGAGCAGCAGUUGCCCCAGCAACUGGAGUUACUCAUGAGUCUGGUUCGCGAUGCCUUUUUGAAUGC .....(((((((((((((((((((((((((((.((......)).)))))))))))))..(((((((....)))))))))).))))))...)))))(((((.((.....)).))))).. ( -41.60) >DroVir_CAF1 116 118 - 1 AUUUGCAGAUUGAGUGCCUGUUCUAUGUUUUGACCUGCAUUGGCCAGGAUAUGGAGCAGCAGCUGCCGCAGCAGCUGGAGCUGCUCAUGUCCCUGGUGCGCGAUGCGUUCCUCAAUGC ....((((((((((((((((((((((((((((.((......)).)))))))))))))))(((((((....)))))))..)).))))).)))...((.((((...)))).))....))) ( -48.70) >DroGri_CAF1 116 118 - 1 AUAUGAAGAUUGAGUGCCUGUUUUACGUUUUGACCUGCAUUGGACAGGACAUGGAGCAACAGCUGCCACAGCAAUUGGAGCUGCUCAUGUCCUUGGUGCGCGAUGCGUUUCUCAAUGC ........((((((..........((((.(((....((((..(...(((((((.((((...(((.(((.......))))))))))))))))))..)))).))).))))..)))))).. ( -35.70) >DroMoj_CAF1 116 118 - 1 UUUUAUAGAUUGAGUCCUUAUUCUAUGUACUGACCUGCAUUGGUCAGGAUAUGGAGCAGCAGCUGCCGCAGCAGUUGGAGCAACUGAUGUCCGUGGUGCGCGAUGCGUUCCUCAAUGC ........((((((.......(((((((.((((((......)))))).)))))))(((...((((((((..((((((...))))))......)))))).))..)))....)))))).. ( -40.80) >DroAna_CAF1 332 118 - 1 CCCUUUAGAUUGAGUGCCUUUUCUAUGUGCUCACCUGCAUCGGUCAGGAUAUGGAGCAGCAGCUGCCACAACAAUUGGAGCUACUUAUGGGCCUCGUUCGUGAUGCUUUUCUGAAUGC ...........(((.((((.((((((((.((.(((......))).)).)))))))).((.((((.(((.......))))))).))...)))))))(((((.((......))))))).. ( -32.20) >DroPer_CAF1 2939 118 - 1 CGUUCCAGGUUGAGUGUCUGUUCUAUGUUCUAACCUGCAUUGGCCAGGACGUGGAGCAGCAGCUGCCCCAGCAGCUGGAGCUGCUCAUGAGCCUCGUGCGCGAUGCCUUCUUGAAUGC (((((.((((...((((((((((((((((((..((......))..))))))))))))))(((((((....)))))))(((..(((....))))))..))))...))))....))))). ( -48.50) >consensus AUUUGCAGAUUGAGUGCCUGUUCUAUGUUCUGACCUGCAUUGGCCAGGACAUGGAGCAGCAGCUGCCACAGCAACUGGAGCUACUCAUGUCCCUGGUGCGCGAUGCGUUCCUCAAUGC ..........((((((((((((((((((((((.((......)).)))))))))))))))(((((((....)))))))..)).)))))............................... (-29.42 = -29.57 + 0.14)

| Location | 9,903,871 – 9,903,973 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -20.53 |
| Energy contribution | -21.03 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.846646 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9903871 102 + 22407834 CCAGUUGCUGGGGCAACUGCUGCUCCAUGUCCUGGCCAAUACAGGUCAAAACAUAAAACAAGCACUCAAUCUGAAAGUU-AUGAAU------AGUAA----UCACAUGUAUUA- .(((.((((((((((.....))))))((((..(((((......)))))..))))......))))......)))...(((-((....------.))))----)...........- ( -22.70) >DroGri_CAF1 156 106 + 1 CCAAUUGCUGUGGCAGCUGUUGCUCCAUGUCCUGUCCAAUGCAGGUCAAAACGUAAAACAGGCACUCAAUCUUCAUAUGCAAGAAUUA----AAUAA----AUAAAUGGAAUAC (((..((((((((.(((....)))))))(.(((((.....))))).).............)))).....(((((....).))))....----.....----.....)))..... ( -21.50) >DroSec_CAF1 156 101 + 1 CCAGUUGCUGGGGCAGUUGCUGCUCCAUGUCCUGGCCAAUACAGGUCAGUACAUAGAACAGGCACUCAAUCUGCAUGUU-AAUAAU------AGUAA----UUA-AUGUUUUA- ..((((((((((((((...)))))))((((.((((((......)))))).))))..((((.(((.......))).))))-......------)))))----)).-........- ( -31.80) >DroSim_CAF1 156 102 + 1 CCAGUUGCUGGGGCAGCUGCUGCUCCAUGUCCUGGCCAAUACAGGUCAGUACAUAGAACAGGCACUCAAUCUGAAUGUU-AAGAAU------GGUAA----UCAUAUGUAUUA- .(((((((....)))))))(((.((.((((.((((((......)))))).)))).)).)))..((.((.(((.......-.))).)------)))..----............- ( -30.40) >DroEre_CAF1 156 111 + 1 CCAGUUGCUGGGGCAGCUGCUGCUCCAUGUCCUGGCCAAUACAGGUUAGUACAUAGAACAGGCACUCAAUCUGAAAAUU-AAGAAU-AUAAUCGUAGUUAUGUAUGUAUAUUA- .(((((((....)))))))(((.((.((((.((((((......)))))).)))).)).)))(((.....(((.......-.)))((-(((((....))))))).)))......- ( -29.40) >DroYak_CAF1 156 107 + 1 CCAGUUGCUGUGGCAGCUGCUGUUCCAUGUCCUGGCCAAUACAAGUCAGUACAUAGAACAGGCACUCAAUCUGAAAGUU-AAGAUU-ACAAUAAUAA----GUAUGUAUAUUA- .(((((((....)))))))((((((.((((.(((((........))))).)))).))))))......(((((.......-.)))))-..........----............- ( -30.50) >consensus CCAGUUGCUGGGGCAGCUGCUGCUCCAUGUCCUGGCCAAUACAGGUCAGUACAUAGAACAGGCACUCAAUCUGAAAGUU_AAGAAU______AGUAA____UUAUAUGUAUUA_ .....(((((((((((...)))))))((((.((((((......)))))).))))......)))).................................................. (-20.53 = -21.03 + 0.50)


| Location | 9,903,871 – 9,903,973 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 81.53 |
| Mean single sequence MFE | -27.98 |
| Consensus MFE | -23.15 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
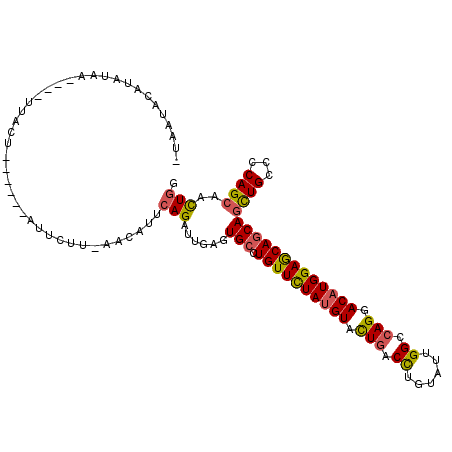
>2L_DroMel_CAF1 9903871 102 - 22407834 -UAAUACAUGUGA----UUACU------AUUCAU-AACUUUCAGAUUGAGUGCUUGUUUUAUGUUUUGACCUGUAUUGGCCAGGACAUGGAGCAGCAGUUGCCCCAGCAACUGG -.......(((((----.....------..))))-)((((.......))))..(((((((((((((((.((......)).)))))))))))))))(((((((....))))))). ( -31.50) >DroGri_CAF1 156 106 - 1 GUAUUCCAUUUAU----UUAUU----UAAUUCUUGCAUAUGAAGAUUGAGUGCCUGUUUUACGUUUUGACCUGCAUUGGACAGGACAUGGAGCAACAGCUGCCACAGCAAUUGG ...((((((....----.((((----((((..((......))..))))))))(((((((...((........))...)))))))..))))))(((..((((...))))..))). ( -22.50) >DroSec_CAF1 156 101 - 1 -UAAAACAU-UAA----UUACU------AUUAUU-AACAUGCAGAUUGAGUGCCUGUUCUAUGUACUGACCUGUAUUGGCCAGGACAUGGAGCAGCAACUGCCCCAGCAACUGG -........-...----.....------......-.....((((.(((.....(((((((((((.(((.((......)).))).)))))))))))))))))).((((...)))) ( -28.70) >DroSim_CAF1 156 102 - 1 -UAAUACAUAUGA----UUACC------AUUCUU-AACAUUCAGAUUGAGUGCCUGUUCUAUGUACUGACCUGUAUUGGCCAGGACAUGGAGCAGCAGCUGCCCCAGCAACUGG -............----.....------......-..(((((.....))))).(((((((((((.(((.((......)).))).)))))))))))(((.(((....))).))). ( -30.60) >DroEre_CAF1 156 111 - 1 -UAAUAUACAUACAUAACUACGAUUAU-AUUCUU-AAUUUUCAGAUUGAGUGCCUGUUCUAUGUACUAACCUGUAUUGGCCAGGACAUGGAGCAGCAGCUGCCCCAGCAACUGG -..........................-...(((-((((....)))))))...(((((((((((.((..((......))..)).)))))))))))(((.(((....))).))). ( -26.80) >DroYak_CAF1 156 107 - 1 -UAAUAUACAUAC----UUAUUAUUGU-AAUCUU-AACUUUCAGAUUGAGUGCCUGUUCUAUGUACUGACUUGUAUUGGCCAGGACAUGGAACAGCAGCUGCCACAGCAACUGG -.....((((...----.......)))-).....-......(((......(((.((((((((((.(((.((......)).))).)))))))))))))((((...))))..))). ( -27.80) >consensus _UAAUACAUAUAA____UUACU______AUUCUU_AACAUUCAGAUUGAGUGCCUGUUCUAUGUACUGACCUGUAUUGGCCAGGACAUGGAGCAGCAGCUGCCCCAGCAACUGG .........................................(((......(((.((((((((((.(((.((......)).))).)))))))))))))((((...))))..))). (-23.15 = -22.98 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:08 2006