
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,887,417 – 9,887,620 |
| Length | 203 |
| Max. P | 0.925176 |

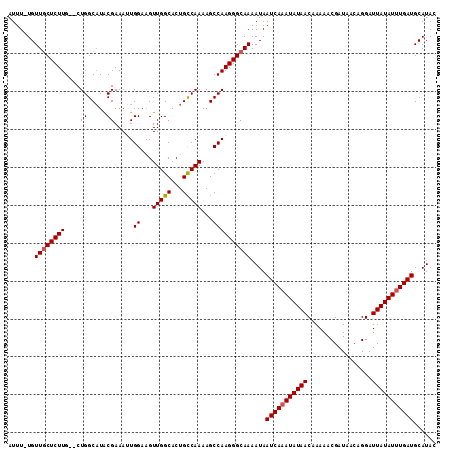
| Location | 9,887,417 – 9,887,529 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -20.03 |
| Energy contribution | -20.19 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9887417 112 + 22407834 AUUUCUGUUGCUCUUGCGCUGGCAUACGAAAUUGGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUA- .......(((((((((.(((((((.....(((.....))).....))))...))))))))))))....(((((((((((.................))))))))))).....- ( -26.83) >DroSec_CAF1 95289 110 + 1 AUUU-UGUUGCUCUUG--CUGGCAUGCGAAAUUGGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUAC ....-..(((((((((--(((((((((.((........)).))).))))...)).)))))))))....(((((((((((.................)))))))))))...... ( -28.43) >DroSim_CAF1 102107 110 + 1 AUUU-UGUUGCUCUUG--CUGGCAUGCGAAAUUGGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUAC ....-..(((((((((--(((((((((.((........)).))).))))...)).)))))))))....(((((((((((.................)))))))))))...... ( -28.43) >DroEre_CAF1 100267 109 + 1 AUUU-UGUUGCUCUUG--CUGGCUUACGAAA-UGGAAGUUGGCACUGUCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUAC ....-..(((((((((--(..((((.(....-..)))))..)).((......))..))))))))....(((((((((((.................)))))))))))...... ( -23.93) >DroYak_CAF1 101387 109 + 1 AUUU-UGUUACUCUUG--CUGGCAUACGAAA-UGGAUGUUGGCACUGUCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUAUGAUGCAUAC .(((-(((((....((--(..((((.(....-..)))))..))).........(((...)))..............)))))))).........(.((((....)))).).... ( -19.10) >consensus AUUU_UGUUGCUCUUG__CUGGCAUACGAAAUUGGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUAC .......((((((((....((.....)).....((...(((((...)))))...))))))))))....(((((((((((.................)))))))))))...... (-20.03 = -20.19 + 0.16)


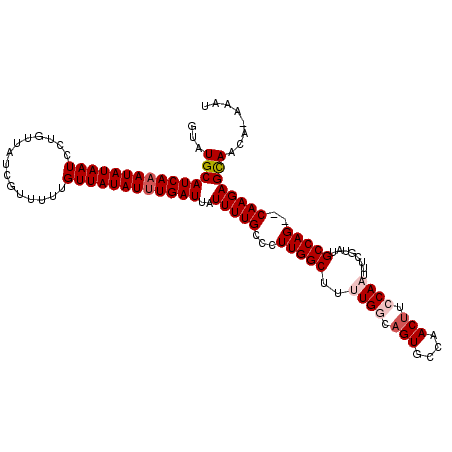
| Location | 9,887,417 – 9,887,529 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 95.50 |
| Mean single sequence MFE | -24.76 |
| Consensus MFE | -19.46 |
| Energy contribution | -20.50 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818186 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9887417 112 - 22407834 -UAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCCAAUUUCGUAUGCCAGCGCAAGAGCAACAGAAAU -..(((((((((((((((...............))))))))))))..((((((.((.(((..((((.(((....))).))))........))))).)))))))))........ ( -26.86) >DroSec_CAF1 95289 110 - 1 GUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCCAAUUUCGCAUGCCAG--CAAGAGCAACA-AAAU ((.(((((((((((((((...............))))))))))))..((((((...((((..((((.(((....))).))))........)))))--)))))))))).-.... ( -25.76) >DroSim_CAF1 102107 110 - 1 GUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCCAAUUUCGCAUGCCAG--CAAGAGCAACA-AAAU ((.(((((((((((((((...............))))))))))))..((((((...((((..((((.(((....))).))))........)))))--)))))))))).-.... ( -25.76) >DroEre_CAF1 100267 109 - 1 GUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGACAGUGCCAACUUCCA-UUUCGUAAGCCAG--CAAGAGCAACA-AAAU ((.(((((((((((((((...............))))))))))))..((((((...((((((.(((.((((........))-))))).)))))))--)))))))))).-.... ( -26.66) >DroYak_CAF1 101387 109 - 1 GUAUGCAUCAUAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGACAGUGCCAACAUCCA-UUUCGUAUGCCAG--CAAGAGUAACA-AAAU ((.(((((((.(((((((...............))))))).))))..((((((...((((...(((.((((........))-)))))...)))))--)))))))))).-.... ( -18.76) >consensus GUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCCAAUUUCGUAUGCCAG__CAAGAGCAACA_AAAU ...(((((((((((((((...............))))))))))))..(((((...(((((..((((.(((....))).))))........)))))..))))))))........ (-19.46 = -20.50 + 1.04)

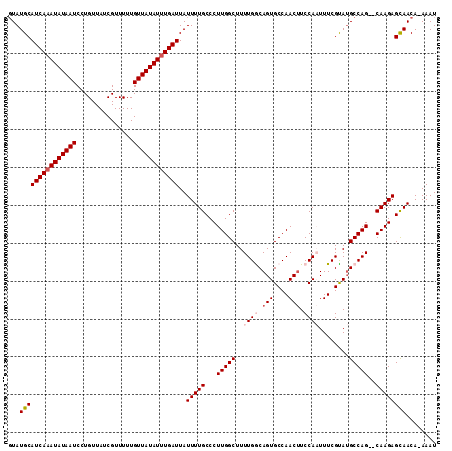
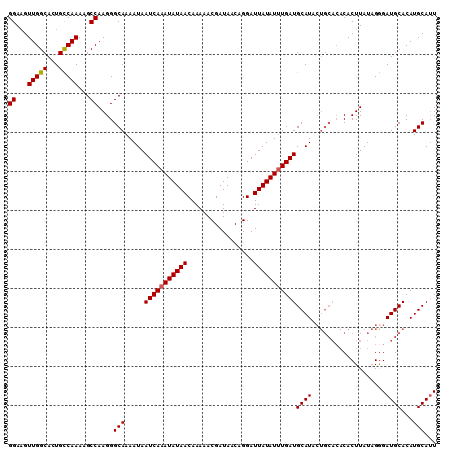
| Location | 9,887,450 – 9,887,553 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -21.34 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.60 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.925176 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9887450 103 + 22407834 GGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUA-------CACUUAUAGGGAUGCACAUGCAAU ((...(((((...)))))...))....(((.....(((((((((((.................)))))))))))((((.-------..((....)).))))...)))... ( -22.53) >DroSec_CAF1 95319 110 + 1 GGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUACUGCACACACUUAUAGGGAUGCACAUGCAUU ((...(((((...)))))...))............(((((((((((.................)))))))))))((((..((((..(........)..)))).))))... ( -23.23) >DroSim_CAF1 102137 110 + 1 GGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUACUGCACACACUUAUAGGGAUGCACAUGCAUU ((...(((((...)))))...))............(((((((((((.................)))))))))))((((..((((..(........)..)))).))))... ( -23.23) >DroEre_CAF1 100296 110 + 1 GGAAGUUGGCACUGUCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUACUGCACACACUUAUAGAGAUGCACAUGCAUU ..(((((((((.((((....(((...)))......(((((((((((.................)))))))))))).))).))).)).)))).....(((((....))))) ( -19.83) >DroYak_CAF1 101416 110 + 1 GGAUGUUGGCACUGUCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUAUGAUGCAUACUGCACACACUUAUAGGGAUGCACAUGCAUU ((...(((((...)))))...)).........((((((...(((..........)))....))))))....(((((((..((((..(........)..)))).))))))) ( -17.90) >consensus GGAAGUUGGCACUGCCAAAAGCCAAGGGCAAAAUAAUCAAAUAUAACAAAAACGAUAACAGGAUUAUAUUUGAUGCAUACUGCACACACUUAUAGGGAUGCACAUGCAUU ((...(((((...)))))...))....(((.....(((((((((((.................)))))))))))((((...................))))...)))... (-18.64 = -18.60 + -0.04)

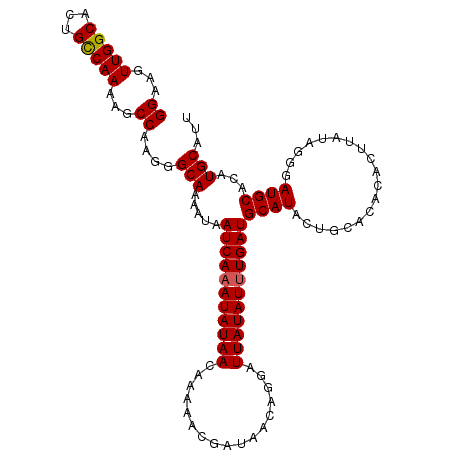
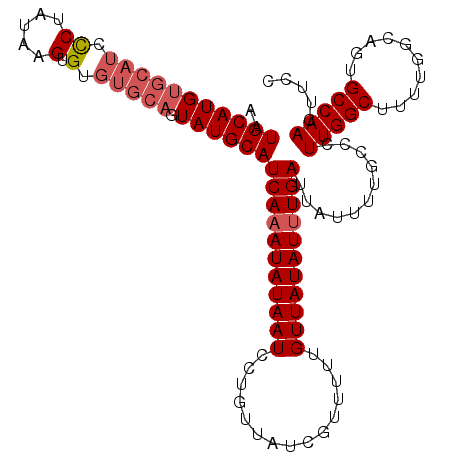
| Location | 9,887,450 – 9,887,553 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 95.45 |
| Mean single sequence MFE | -22.86 |
| Consensus MFE | -19.38 |
| Energy contribution | -20.42 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.735163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9887450 103 - 22407834 AUUGCAUGUGCAUCCCUAUAAGUG-------UAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCC ..((((((..(..........)..-------))))))(((((((((((...............))))))))))).......(((...)))..(((((...)))))..... ( -21.76) >DroSec_CAF1 95319 110 - 1 AAUGCAUGUGCAUCCCUAUAAGUGUGUGCAGUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCC ..((((((((((..((.....).)..)))).))))))(((((((((((...............))))))))))).......(((...)))..(((((...)))))..... ( -24.66) >DroSim_CAF1 102137 110 - 1 AAUGCAUGUGCAUCCCUAUAAGUGUGUGCAGUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCC ..((((((((((..((.....).)..)))).))))))(((((((((((...............))))))))))).......(((...)))..(((((...)))))..... ( -24.66) >DroEre_CAF1 100296 110 - 1 AAUGCAUGUGCAUCUCUAUAAGUGUGUGCAGUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGACAGUGCCAACUUCC ..((((((((((((.(.....).).))))).))))))(((((((((((...............)))))))))))...........(((((..........)))))..... ( -22.96) >DroYak_CAF1 101416 110 - 1 AAUGCAUGUGCAUCCCUAUAAGUGUGUGCAGUAUGCAUCAUAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGACAGUGCCAACAUCC ..((((((((((..((.....).)..)))).))))))(((.(((((((...............))))))).)))...........(((((..........)))))..... ( -20.26) >consensus AAUGCAUGUGCAUCCCUAUAAGUGUGUGCAGUAUGCAUCAAAUAUAAUCCUGUUAUCGUUUUUGUUAUAUUUGAUUAUUUUGCCCUUGGCUUUUGGCAGUGCCAACUUCC ..(((((((((((.((.....).).))))).))))))(((((((((((...............)))))))))))...........(((((..........)))))..... (-19.38 = -20.42 + 1.04)


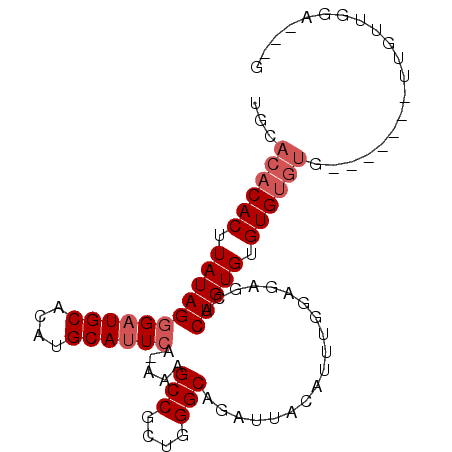
| Location | 9,887,529 – 9,887,620 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -26.96 |
| Consensus MFE | -21.04 |
| Energy contribution | -22.04 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9887529 91 + 22407834 ------CACUUAUAGGGAUGCACAUGCAAUCAUAAAGCCGCUGGGCAGAUUACAUUUGGAGAGCACUGUGUGUGUGUGUGUGUUGUUGUUGUAGCCG ------.((..((((.((((((((((((........(((....)))....(((((.((.....))..))))))))))))))))).)))).))..... ( -23.70) >DroSec_CAF1 95399 86 + 1 UGCACACACUUAUAGGGAUGCACAUGCAUUCA-AAAGCCGCUGGGCAGAUUACAUUUGGAGAGCACUGUGUGUGUGUG-------UUGUUGGA---G .((((((((.(((((((((((....)))))).-...(((....)))...................))))).)))))))-------).......---. ( -28.70) >DroSim_CAF1 102217 86 + 1 UGCACACACUUAUAGGGAUGCACAUGCAUUCA-AAAGCCGCUGGGCAGAUUACAUUUGGAGAGCACUGUGUGUGUGUG-------UUGUUGGA---G .((((((((.(((((((((((....)))))).-...(((....)))...................))))).)))))))-------).......---. ( -28.70) >DroEre_CAF1 100376 85 + 1 UGCACACACUUAUAGAGAUGCACAUGCAUUCA-AAAGCCGCUGGGCAGAUUACAUUUGAGAAACACUGUGUGUGUGUA-------G-CUCAGA---A .((((((((.(((((.(((((....)))))..-...(((....)))...................))))).)))))).-------)-).....---. ( -23.70) >DroYak_CAF1 101496 85 + 1 UGCACACACUUAUAGGGAUGCACAUGCAUUCG-AAAGCCGCUGGGCAGAUUACAUUUGUGGAGCACUGUGUGUGUGUA-------G-CUUGGA---A .((((((((.(((((((((((....)))))).-......(((..((((((...))))))..))).))))).)))))).-------)-).....---. ( -30.00) >consensus UGCACACACUUAUAGGGAUGCACAUGCAUUCA_AAAGCCGCUGGGCAGAUUACAUUUGGAGAGCACUGUGUGUGUGUG_______UUGUUGGA___G ...((((((.(((((((((((....)))))).....(((....)))...................))))).)))))).................... (-21.04 = -22.04 + 1.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:52:04 2006