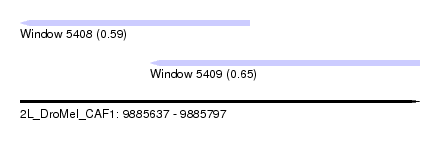
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,885,637 – 9,885,797 |
| Length | 160 |
| Max. P | 0.647271 |

| Location | 9,885,637 – 9,885,729 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 84.89 |
| Mean single sequence MFE | -12.59 |
| Consensus MFE | -9.44 |
| Energy contribution | -9.68 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586030 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9885637 92 - 22407834 CAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUCUUCCCAAGCGCUUUUCCAACUUUCCGCUUUCCU-CCCG-AAAUUUUGCCCACAAAAA ...((((((.(((..((((((......))))))..)))....(((((..............)))))....-....-....))))))........ ( -16.04) >DroSec_CAF1 93536 90 - 1 CAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUC--CCCAAGCGCUUUUCCACUUUUCCGCUUUCCC-CUC-CCCGAUUUCCCCACAAAAA ..(((..........((((((......))))))....--...(((((..............)))))...)-)).-................... ( -11.44) >DroSim_CAF1 100388 91 - 1 CAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUC--CCCAAGCGCUUUUCCACUUUUCCGCUUUCCC-CCCGCCCAAUUUCCCCACAAAAA ...(((.........((((((......))))))....--...(((((..............)))))....-...)))................. ( -12.54) >DroEre_CAF1 98454 81 - 1 CAAGGUAAACAGAACGAGCAA-AAUUGUUGCUCUCUU--CCCCAGCGCUUUUCCACUUUUCUGCUU--------UCCCCAUUUCCCAACA--AA ...((.((((((((.((((((-.....))))))....--....((.(......).)).))))).))--------).))............--.. ( -11.30) >DroYak_CAF1 99552 90 - 1 CAAGGUAAACAGAACGAGCCA-AAUUGUUGCUCUCUC--CCCAAGCGCUUUUCCACUUUUCCGCUUCUCCGCUAUCCCCAUUUACCCACAA-AA ...((((((......((((..-.......))))....--...(((((..............)))))..............)))))).....-.. ( -11.64) >consensus CAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUC__CCCAAGCGCUUUUCCACUUUUCCGCUUUCCC_CCCGCCCAAUUUCCCCACAAAAA ...((.....(((..((((((......)))))))))....))(((((..............)))))............................ ( -9.44 = -9.68 + 0.24)

| Location | 9,885,689 – 9,885,797 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Mean single sequence MFE | -17.73 |
| Consensus MFE | -11.67 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.647271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
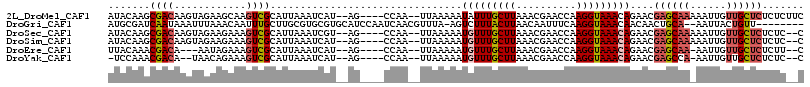
>2L_DroMel_CAF1 9885689 108 - 22407834 AUACAAGCGACAAGUAGAAGCAAGUCGCAUUAAAUCAU--AG----CCAA--UUAAAAAUAUUUGCUUAAACGAACCAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUCUUC .(((..(((((..((....))..)))))..........--((----(.((--(.......))).))).............)))...(((..((((((......))))))..))).. ( -19.00) >DroGri_CAF1 106379 105 - 1 AUGCGAUCAAUAAAUUUAAACAAUUUGCUUGCGUGCGUGCAUCCAAUCAACGUUUA-AGUCUUUACUUAACAAUUUCAAGGUAAACAACAACUGCA--AAUUACUGUU-------- .((((........((((((((...(((.(((.(((....))).))).))).)))))-))).(((((((..........))))))).......))))--..........-------- ( -14.20) >DroSec_CAF1 93588 106 - 1 AUACAAGCGACAAGUAGAAGAAAGUCGCAUUAAAUCGU--AG----CCAA--UUAAAAAUGUUUGCUUAAACGAACCAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUC--C ......(((((............)))))..........--..----....--.......(((((((((..........)))))))))....((((((......))))))....--. ( -21.30) >DroSim_CAF1 100441 106 - 1 AUACAAGCGACAAGUAGAAGAAAGUCGCAUUAAAUCAU--AG----CCAA--UUAAAAAUGUUUGCUUAAACGAACCAAGGUAAACAGAACGAGCAAAAAUUGUUGCUCUCUC--C ......(((((............)))))..........--..----....--.......(((((((((..........)))))))))....((((((......))))))....--. ( -21.30) >DroEre_CAF1 98498 102 - 1 UUACAAACGACA---AAUAGAAAGUCGCAUUAAAUCAU--AG----CCAA--UUAAAAAUGUUUGCUUAAACGAACCAAGGUAAACAGAACGAGCAA-AAUUGUUGCUCUCUU--C .......((((.---........))))...........--..----....--.......(((((((((..........)))))))))....((((((-.....))))))....--. ( -17.00) >DroYak_CAF1 99605 102 - 1 -UCCAAACGACA--UAACAGAAAGUCGCAUUAAAUCAU--AG----CCAA--UUAAAAAUGUUUGCUUAAACGAACCAAGGUAAACAGAACGAGCCA-AAUUGUUGCUCUCUC--C -......((((.--.........))))...........--..----....--.......(((((((((..........)))))))))....((((..-.......))))....--. ( -13.60) >consensus AUACAAGCGACAAGUAGAAGAAAGUCGCAUUAAAUCAU__AG____CCAA__UUAAAAAUGUUUGCUUAAACGAACCAAGGUAAACAGAACGAGCAA_AAUUGUUGCUCUCUC__C .......((((............))))................................(((((((((..........)))))))))....((((((......))))))....... (-11.67 = -12.28 + 0.61)
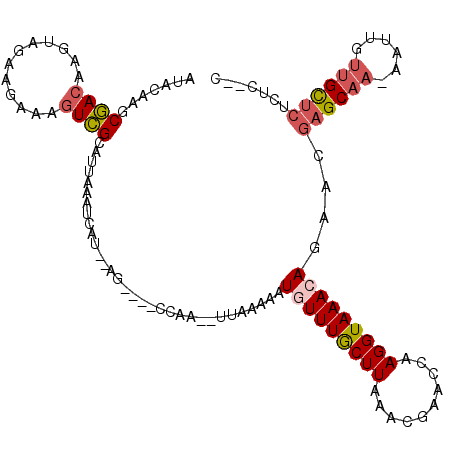
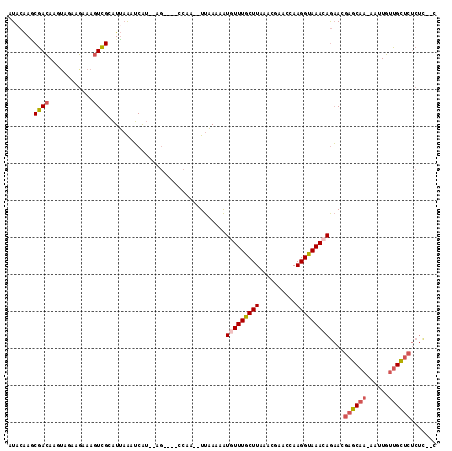
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:59 2006