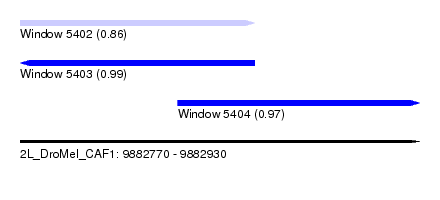
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,882,770 – 9,882,930 |
| Length | 160 |
| Max. P | 0.985944 |

| Location | 9,882,770 – 9,882,864 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -28.31 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.860667 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
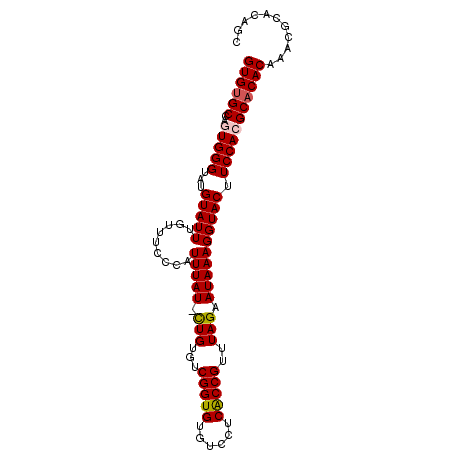
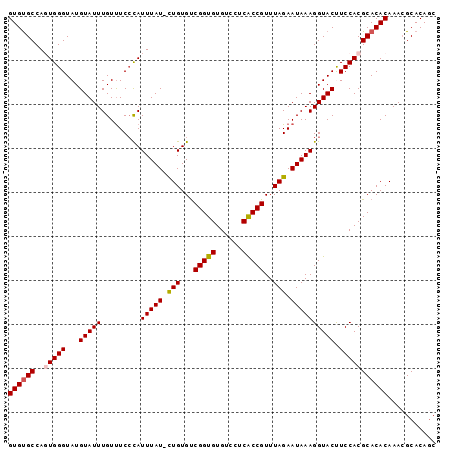
>2L_DroMel_CAF1 9882770 94 + 22407834 CACAAACCAGUGUUGUACACACGCAAAGUGGAGCAAUUCCGACAACUCCAUUUGCAUCGGUAUGUGUGCCAGUGGGUAUGUAUUUGUUUCCCAU ((((.(((.((((.....))))(((((.(((((............))))))))))...))).)))).....(((((.............))))) ( -26.62) >DroSec_CAF1 90669 93 + 1 CACACACCAGUGU-AUACAUACGCAAAGUGGAGCAAUUCCGACAACUCCAUUUGCAUCGGUAUGUGCGCCACUGGGUAUGUAUUUGUUUCCCAU .((((....))))-((((((((.((..((((.(((...(((((((......)))..))))....))).)))))).))))))))........... ( -28.70) >DroSim_CAF1 97522 93 + 1 CACACACCAGUGU-AUACAUACGCAAAGUGGAGCAAUUCCGACAACUCCAUUUGUAUCGGUAUGUGUGCCACUGGGUAUGUAUUUGUUUCCCAU .(((..(((((((-(((((((((((((.(((((............))))))))))....))))))))).))))))...)))............. ( -29.60) >consensus CACACACCAGUGU_AUACAUACGCAAAGUGGAGCAAUUCCGACAACUCCAUUUGCAUCGGUAUGUGUGCCACUGGGUAUGUAUUUGUUUCCCAU .(((..((((((..(((((((((((((.(((((............))))))))))....))))))))..))))))...)))............. (-24.16 = -24.83 + 0.67)


| Location | 9,882,770 – 9,882,864 |
|---|---|
| Length | 94 |
| Sequences | 3 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 95.02 |
| Mean single sequence MFE | -31.01 |
| Consensus MFE | -29.59 |
| Energy contribution | -30.37 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.02 |
| SVM RNA-class probability | 0.985944 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9882770 94 - 22407834 AUGGGAAACAAAUACAUACCCACUGGCACACAUACCGAUGCAAAUGGAGUUGUCGGAAUUGCUCCACUUUGCGUGUGUACAACACUGGUUUGUG .((((.............))))......((((.((((..(((((((((((..........)))))).)))))((((.....)))))))).)))) ( -28.22) >DroSec_CAF1 90669 93 - 1 AUGGGAAACAAAUACAUACCCAGUGGCGCACAUACCGAUGCAAAUGGAGUUGUCGGAAUUGCUCCACUUUGCGUAUGUAU-ACACUGGUGUGUG ...(....).....(((((((((((....((((((....(((((((((((..........)))))).)))))))))))..-.)))))).))))) ( -34.30) >DroSim_CAF1 97522 93 - 1 AUGGGAAACAAAUACAUACCCAGUGGCACACAUACCGAUACAAAUGGAGUUGUCGGAAUUGCUCCACUUUGCGUAUGUAU-ACACUGGUGUGUG ...(....).....(((((((((((....(((((((((......((((((..........))))))..))).))))))..-.)))))).))))) ( -30.50) >consensus AUGGGAAACAAAUACAUACCCAGUGGCACACAUACCGAUGCAAAUGGAGUUGUCGGAAUUGCUCCACUUUGCGUAUGUAU_ACACUGGUGUGUG ...(....)...((((((((.((((....((((((....(((((((((((..........)))))).)))))))))))....)))))))))))) (-29.59 = -30.37 + 0.78)

| Location | 9,882,833 – 9,882,930 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 93.98 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -24.88 |
| Energy contribution | -25.08 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.966000 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9882833 97 + 22407834 GUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAUUCUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAACGCACAGC ((((((..(((((...(((........((.(((((((((...(((((......)))))..)))))))))))))).)))))))))))........... ( -33.00) >DroSec_CAF1 90731 96 + 1 GUGCGCCACUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGCCGGUGUGUCCUCGCCGUUUAGAAUAAAGGUACCUCCACGCACACAAACGCACAGC (((((....((((.............)))).....-.((((((((((......)))))...........((.....))..)))))....)))))... ( -25.82) >DroSim_CAF1 97584 96 + 1 GUGUGCCACUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGUCGGUGUGUCCUCGCCGUUUAGAAUAAAGGUACCUCCACGCACACAAACGCACAGC ((((((....((((((...(((((((..(((....-..))).(((((......)))))....))))))).))))))....))))))........... ( -25.90) >DroEre_CAF1 95705 96 + 1 GUGUGCCAGUGGGUAUGUAUUUGUCUCGUAUUUAU-UUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAAGGCACAGC ((((((..(((((...(((((.........(((((-((....(((((......)))))....)))))))))))).)))))))))))........... ( -28.90) >DroYak_CAF1 95202 96 + 1 GUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAU-CUGUGUCGGUGUGCCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAACGCACAUC ((((((..(((((...((((((..(((.(((....-..))).(((((......)))))....)))...)))))).)))))))))))........... ( -28.40) >consensus GUGUGCCAGUGGGUAUGUAUUUGUUUCCCAUUUAU_CUGUGUCGGUGUGUCCUCACCGUUUAGAAUAAAGGUACUUCCACGCACACAAACGCACAGC ((((((..(((((...(((((.........(((((.(((...(((((......)))))..))).)))))))))).)))))))))))........... (-24.88 = -25.08 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:55 2006