
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,877,250 – 9,877,374 |
| Length | 124 |
| Max. P | 0.966921 |

| Location | 9,877,250 – 9,877,347 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 86.70 |
| Mean single sequence MFE | -26.92 |
| Consensus MFE | -18.88 |
| Energy contribution | -18.28 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.26 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.966921 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9877250 97 + 22407834 GUUGCUGCAGCUGCAAAGUAUCGGCGACCUUUUGGCUUUUACAAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAU (((((((..(((....)))..)))))))...............((((((((..(((((((....))))).))..))))))))............... ( -25.50) >DroGri_CAF1 95801 97 + 1 GUGGCUGCAGCUGCAAGCAAGCGGCGGCCUUAUCGUUUUGACAAUGGAACAAACACAACAAAAAUGUUGAGGAAUGUUCCAUAAUUAAUUAAGAAUU ..((((((.(((.......))).))))))...((...((((..((((((((..(.(((((....))))).)...))))))))..))))....))... ( -31.30) >DroSim_CAF1 92189 97 + 1 GUUGCUGCAGCUGCAAAGUAUCGGCGACCUUUUGGCUUUUACAAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAU (((((((..(((....)))..)))))))...............((((((((..(((((((....))))).))..))))))))............... ( -25.50) >DroYak_CAF1 89589 97 + 1 GUUGCUGCAGCUGCAAACAAUCGGCGACCUUUUUGCUUUUACAAUGGAACAAACACAACGAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAU (((((((.(..((....)).))))))))...............((((((((..(((((((....))))).))..))))))))............... ( -21.00) >DroMoj_CAF1 135670 97 + 1 UUGGCUGCAGCUGGAAGCGGGCGACGGCCUUUUCGUUUUGGCAAUGGAACAAACACAACAAAUAUGUUGAUGAAUGUUCCAUAAUUAAUUGAGAAAU ..(((((..(((.(...).)))..)))))((((((..((((..((((((((..(((((((....))))).))..))))))))..)))).)))))).. ( -31.30) >consensus GUUGCUGCAGCUGCAAACAAUCGGCGACCUUUUCGCUUUUACAAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAU (..((....))..)........(((((.....)))))......((((((((..(((((((....))))).))..))))))))............... (-18.88 = -18.28 + -0.60)



| Location | 9,877,250 – 9,877,347 |
|---|---|
| Length | 97 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 86.70 |
| Mean single sequence MFE | -24.88 |
| Consensus MFE | -17.60 |
| Energy contribution | -17.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.87 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9877250 97 - 22407834 AUUUUCAUAUUAAUUAUGGAACAUUCAUCAACAAUUUUGUUGUGUUUGUUCCAUUGUAAAAGCCAAAAGGUCGCCGAUACUUUGCAGCUGCAGCAAC ................(((((((..((.(((((....)))))))..)))))))((((...(((...(((((.......)))))...)))...)))). ( -23.20) >DroGri_CAF1 95801 97 - 1 AAUUCUUAAUUAAUUAUGGAACAUUCCUCAACAUUUUUGUUGUGUUUGUUCCAUUGUCAAAACGAUAAGGCCGCCGCUUGCUUGCAGCUGCAGCCAC ................(((((((..(..(((((....))))).)..)))))))(((((.....)))))(((.((.(((.(....)))).)).))).. ( -29.00) >DroSim_CAF1 92189 97 - 1 AUUUUCAUAUUAAUUAUGGAACAUUCAUCAACAAUUUUGUUGUGUUUGUUCCAUUGUAAAAGCCAAAAGGUCGCCGAUACUUUGCAGCUGCAGCAAC ................(((((((..((.(((((....)))))))..)))))))((((...(((...(((((.......)))))...)))...)))). ( -23.20) >DroYak_CAF1 89589 97 - 1 AUUUUCAUAUUAAUUAUGGAACAUUCAUCAACAAUUUCGUUGUGUUUGUUCCAUUGUAAAAGCAAAAAGGUCGCCGAUUGUUUGCAGCUGCAGCAAC .........(((...((((((((..((.((((......))))))..))))))))..)))..(((((...(((...)))..))))).((....))... ( -22.90) >DroMoj_CAF1 135670 97 - 1 AUUUCUCAAUUAAUUAUGGAACAUUCAUCAACAUAUUUGUUGUGUUUGUUCCAUUGCCAAAACGAAAAGGCCGUCGCCCGCUUCCAGCUGCAGCCAA .((((..........((((((((..((.(((((....)))))))..)))))))).........)))).(((.((.((.........)).)).))).. ( -26.11) >consensus AUUUUCAUAUUAAUUAUGGAACAUUCAUCAACAAUUUUGUUGUGUUUGUUCCAUUGUAAAAGCCAAAAGGUCGCCGAUAGUUUGCAGCUGCAGCAAC ...............((((((((..((.(((((....)))))))..))))))))...............((.((.(........).)).))...... (-17.60 = -17.60 + -0.00)

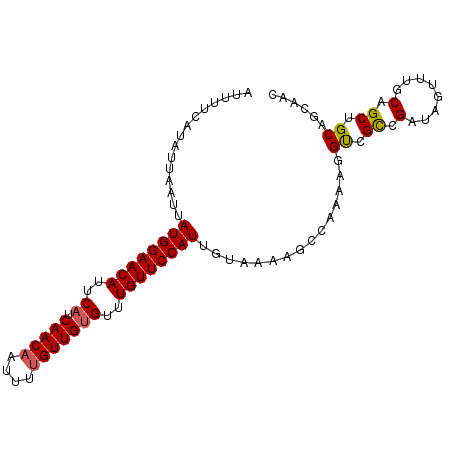
| Location | 9,877,270 – 9,877,374 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 86.73 |
| Mean single sequence MFE | -24.10 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.45 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.75 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9877270 104 + 22407834 UCGGCGACCUUUUGGCUUUUA--C-AAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAUUGUAUUGAAUUCGUUCUCU--UUUCGUUC-------- ..(((((((....))......--.-.((((((((..(((((((....))))).))..))))))))..((((((..(...)..))))))..)))))....--........-------- ( -21.00) >DroSec_CAF1 85320 102 + 1 UCGGCGACCUUUUGGCUUUUA--C-AAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAUUGUAUUGAACUCG--CCCU--UUUCGUUC-------- ..(((((((....))......--.-.((((((((..(((((((....))))).))..))))))))..((((((..(...)..))))))..)))--))..--........-------- ( -25.80) >DroSim_CAF1 92209 102 + 1 UCGGCGACCUUUUGGCUUUUA--C-AAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAUUGUAUUGAACUCG--CUCU--UUUCGUUC-------- ..(((((((....))......--.-.((((((((..(((((((....))))).))..))))))))..((((((..(...)..))))))..)))--))..--........-------- ( -23.40) >DroEre_CAF1 90211 102 + 1 UCGGCGACCUUUUUGCUUUUA--C-AAUGGAACAGACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAUUGUAUUGAACUCG--CUCU--CUUCGUUC-------- ..(((((..............--.-.((((((((..(((((((....))))).))..))))))))..((((((..(...)..))))))..)))--))..--........-------- ( -24.40) >DroYak_CAF1 89609 102 + 1 UCGGCGACCUUUUUGCUUUUA--C-AAUGGAACAAACACAACGAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAUUGUAUUUAACUCG--CUCG--CUUCGUUC-------- .((((((......(((.....--.-.((((((((..(((((((....))))).))..)))))))).......((....)).)))......)))--).))--........-------- ( -19.80) >DroAna_CAF1 93852 115 + 1 UCGGCAACCUUUUCGUUUUGGAACAAAUGGAACAAACUCAACAAAAAUGUUGAUGAAUGUUCCAUAAUUAACAGGAAGAUUUAAUAUAGUUAG--CUCUCUGUUUGUUGCCUUCCCC ..((((((..................((((((((..(((((((....)))))).)..))))))))....(((((..((...((((...)))).--))..))))).))))))...... ( -30.20) >consensus UCGGCGACCUUUUGGCUUUUA__C_AAUGGAACAAACACAACAAAAUUGUUGAUGAAUGUUCCAUAAUUAAUAUGAAAAUUGUAUUGAACUCG__CUCU__UUUCGUUC________ ..(((((...................((((((((..(((((((....))))).))..))))))))..(((((((((...)))))))))...............)))))......... (-18.14 = -18.45 + 0.31)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:50 2006