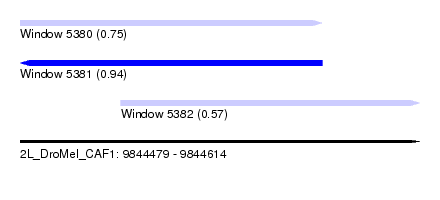
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,844,479 – 9,844,614 |
| Length | 135 |
| Max. P | 0.943510 |

| Location | 9,844,479 – 9,844,581 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -16.05 |
| Energy contribution | -17.61 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.745924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9844479 102 + 22407834 CGGCUUCUG---CCAUC--UCAACUGUGCUGUUACUGACUUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCA ........(---(((((--.....((((.((.((((...((((((((.........))))))))...)))).)).))))....(((....)))......)))))).. ( -30.60) >DroPse_CAF1 56358 89 + 1 --GCUGCUG---C-------------UGCUGCCGCCGGCUUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUCCGCUUCCACCUUAAUUGAUGGCCA --((.((..---.-------------.)).)).((((((((.(((((.........))))))))))..(((....)))........................))).. ( -24.60) >DroSim_CAF1 56204 104 + 1 CGGCUUCUG---CCAUCUUUCAACUGUGCUGUUGCUGACUUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCA ........(---(((((....(((......)))(((((..(((((((.........)))))))(((((((.........))))))).))))).......)))))).. ( -31.50) >DroYak_CAF1 55971 102 + 1 CGGCUUCUG---CCAUC--UCAACUGCGCUGUUGCUGACUUUGAUGCCUUCACUUAGCUUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCA ........(---(((((--......(.((((..((..(..(((.((....(((((.((.....)).)))))....))))).)..))..)))))......)))))).. ( -30.00) >DroAna_CAF1 59816 90 + 1 ---C------------C--UCCGCAAUGCUGUUGCUGACUUUGAUGCCUUUACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCA ---.------------.--.(((((((......(((((..(((((((.........)))))))(((((((.........))))))).)))))....)))).)))... ( -25.70) >DroPer_CAF1 56857 92 + 1 --GCUGCUGCUGC-------------UGCUGCCGCCGGCUUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUCCGCUUCCACCUUAAUUGAUGGCCA --((.((....))-------------.))....((((((((.(((((.........))))))))))..(((....)))........................))).. ( -25.00) >consensus __GCUUCUG___C___C__UCAACUGUGCUGUUGCUGACUUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCA ...........................(((((((((((..(((((((.........)))))))(((((((.........))))))).))))).......)))))).. (-16.05 = -17.61 + 1.56)


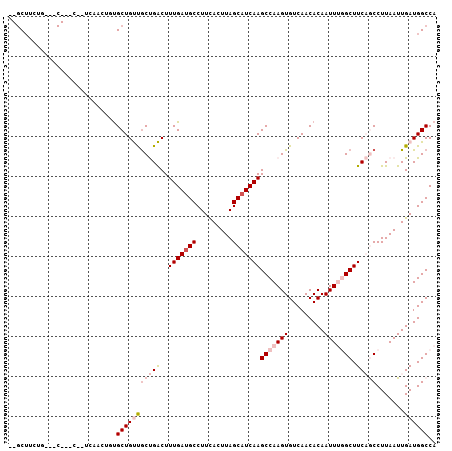
| Location | 9,844,479 – 9,844,581 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 82.31 |
| Mean single sequence MFE | -36.02 |
| Consensus MFE | -23.04 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.22 |
| Mean z-score | -3.82 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.943510 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9844479 102 - 22407834 UGGCCAUCAAUUAAGGCUGAAGCCAAAUUGUGUUGACACUUGGCUUGAUGCUAAGUGAAGGCAUCAAAGUCAGUAACAGCACAGUUGA--GAUGG---CAGAAGCCG ..((((((......(((....))).((((((((((..(((.(((((((((((.......)))))).))))))))..))))))))))..--)))))---)........ ( -42.50) >DroPse_CAF1 56358 89 - 1 UGGCCAUCAAUUAAGGUGGAAGCGGAAUUGUGUUGACACUUGGCUUGAUGCUAAGUGAAGGCAUCAAAGCCGGCGGCAGCA-------------G---CAGCAGC-- ...(((((......)))))...(((..(((((((..((((((((.....))))))))..)))).)))..)))((.((....-------------)---).))...-- ( -31.80) >DroSim_CAF1 56204 104 - 1 UGGCCAUCAAUUAAGGCUGAAGCCAAAUUGUGUUGACACUUGGCUUGAUGCUAAGUGAAGGCAUCAAAGUCAGCAACAGCACAGUUGAAAGAUGG---CAGAAGCCG ..((((((......(((....))).((((((((((.((((((((.....))))))))..(((......))).....))))))))))....)))))---)........ ( -41.80) >DroYak_CAF1 55971 102 - 1 UGGCCAUCAAUUAAGGCUGAAGCCAAAUUGUGUUGACACUUGGCUUGAAGCUAAGUGAAGGCAUCAAAGUCAGCAACAGCGCAGUUGA--GAUGG---CAGAAGCCG ..((((((......(((....))).((((((((((.((((((((.....))))))))..(((......))).....))))))))))..--)))))---)........ ( -41.10) >DroAna_CAF1 59816 90 - 1 UGGCCAUCAAUUAAGGCUGAAGCCAAAUUGUGUUGACACUUGGCUUGAUGCUAAGUAAAGGCAUCAAAGUCAGCAACAGCAUUGCGGA--G------------G--- ...((..((((....(((((.(((((...(((....))))))))((((((((.......))))))))..)))))......)))).)).--.------------.--- ( -26.40) >DroPer_CAF1 56857 92 - 1 UGGCCAUCAAUUAAGGUGGAAGCGGAAUUGUGUUGACACUUGGCUUGAUGCUAAGUGAAGGCAUCAAAGCCGGCGGCAGCA-------------GCAGCAGCAGC-- ...(((((......)))))..((((..(((((((..((((((((.....))))))))..)))).)))..)).((.((....-------------)).)).))...-- ( -32.50) >consensus UGGCCAUCAAUUAAGGCUGAAGCCAAAUUGUGUUGACACUUGGCUUGAUGCUAAGUGAAGGCAUCAAAGUCAGCAACAGCACAGUUGA__G___G___CAGAAGC__ .((((.........))))...........((((((....(((((((((((((.......)))))).)))))))...))))))......................... (-23.04 = -22.98 + -0.05)

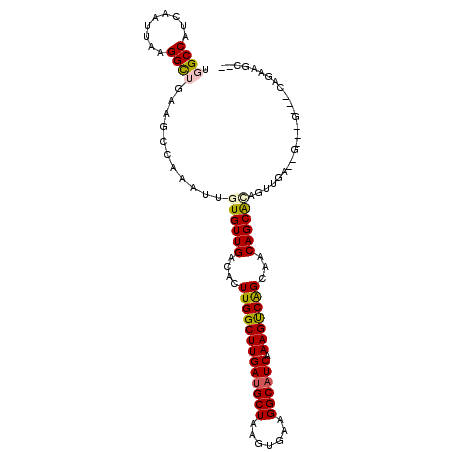
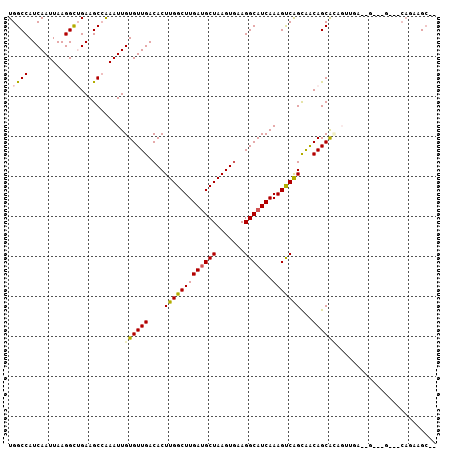
| Location | 9,844,513 – 9,844,614 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 85.23 |
| Mean single sequence MFE | -19.33 |
| Consensus MFE | -14.87 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.570279 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9844513 101 + 22407834 UUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCAUAAUGCACACACACACACAACUCUUCAC---CGGCG---- .(((((((.........)))))))(((..(((....)))....(((((((((......)))).)))))............................---.))).---- ( -23.40) >DroPse_CAF1 56379 100 + 1 UUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUCCGCUUCCACCUUAAUUGAUGGCCAUAAUGCACA--------CAGCUCUGCUUACCCUUCAAGCC .(((((((.........)))))))((...(((....)))...................((((.((...(((.(((..--------......)))))).)).)))))). ( -15.80) >DroEre_CAF1 55807 95 + 1 UUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCAUAAUGCACA------CACAACUCUUCCC---CUCCG---- .(((((((.........)))))))((((.((....)).(((((.(((....)))..))))).))))...........------.............---.....---- ( -20.20) >DroYak_CAF1 56005 91 + 1 UUUGAUGCCUUCACUUAGCUUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCAUAAUGCA----------CAACUCUUCCC---CUGCG---- ......(((.(((.((((((..(((((((((.........))))))))).)))..))).))).)))......(((----------...........---.))).---- ( -20.60) >DroAna_CAF1 59838 96 + 1 UUUGAUGCCUUUACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCAUAAUGCAC--------ACAACUCUUCUUAC-CUGCC---C .(((((((.........)))))))((((.((....)).(((((.(((....)))..))))).))))..........--------..............-.....---. ( -20.20) >DroPer_CAF1 56881 100 + 1 UUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUCCGCUUCCACCUUAAUUGAUGGCCAUAAUGCACA--------CAGCUCUGCUUACCCUUCAAGCC .(((((((.........)))))))((...(((....)))...................((((.((...(((.(((..--------......)))))).)).)))))). ( -15.80) >consensus UUUGAUGCCUUCACUUAGCAUCAAGCCAAGUGUCAACACAAUUUGGCUUCAGCCUUAAUUGAUGGCCAUAAUGCACA________CAACUCUUCUC___CUGCG____ .(((((((.........)))))))((((.((....)).(((((.(((....)))..))))).)))).......................................... (-14.87 = -15.70 + 0.83)

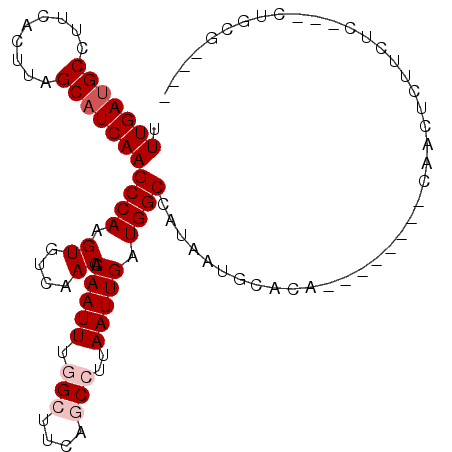
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:34 2006