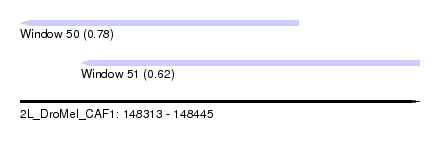
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 148,313 – 148,445 |
| Length | 132 |
| Max. P | 0.778121 |

| Location | 148,313 – 148,405 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.56 |
| Mean single sequence MFE | -25.52 |
| Consensus MFE | -17.72 |
| Energy contribution | -17.28 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.778121 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
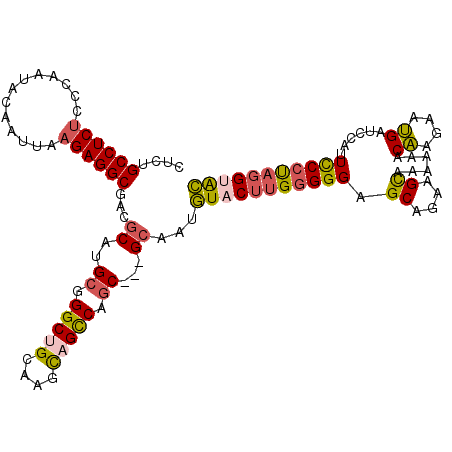
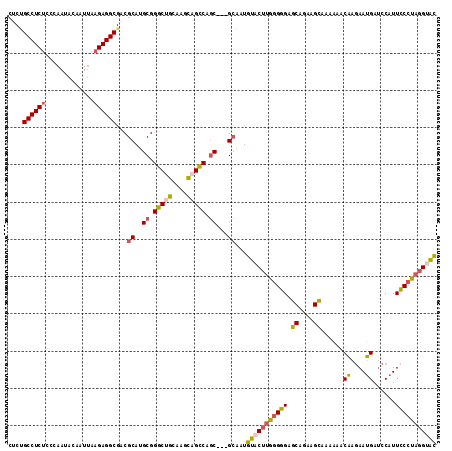
>2L_DroMel_CAF1 148313 92 - 22407834 GCUGCAAGCAGCCAGC---GCAAUGUACUUGGGGGAGCGGAAGCAAAAAACAAGAAUGAUCCAUUCCCCAGGUACGAAACAAGGUUUACU------UAGAG-------------- ((((....))))....---....(((((((((((((((....))......((....)).....)))))))))))))....(((.....))------)....-------------- ( -30.00) >DroSec_CAF1 30757 92 - 1 GCUGCAAGCAGCCAGC---GCAAUGUACUUGGGGGAGCAGAAGCAAAAAACAAGAAUGAUCCAUUCCCUAGGUACGAAACAAGGUUUACC------UAAAG-------------- ((((....))))....---....(((((((((((((((....))......((....)).....))))))))))))).....(((....))------)....-------------- ( -25.90) >DroSim_CAF1 32399 92 - 1 GCUGCAAGCAGCCAGC---GCAAUGUUCUUGGGGGAGCAGAAGCAAAAAACAAGAAUGAUCCAUUCCCUAGGUACGAAACAAGGUUUACC------UAAAG-------------- ((((....))))....---....(((.(((((((((((....))......((....)).....))))))))).))).....(((....))------)....-------------- ( -21.30) >DroEre_CAF1 42371 112 - 1 GCGGCAACCAGCCAGC---GCGAUGUACUUGGGGGCGCGGAGGCUAAACGCAAGAAUGAUCCAUUCCCUAGGUACGAAUAUAGGUUUACCAUUUCCCAAAGGUUCCCAUUUUAAG (((((.....))).))---....((((((((((((...(((..(....(....)...).)))..))))))))))))......((...(((..........)))..))........ ( -30.40) >DroYak_CAF1 30954 91 - 1 GCUGCCAGCAGUCAGC---GCGAUGUACUUGGGGGCGCGGAAGCCAAGCACAAGAAUGAUCCAUUCCCUAGGUACGAA---AGGU------------------UCCCACUUUAAG ((((.(....).))))---....(((((((((((((......)))........(((((...)))))))))))))))((---((..------------------.....))))... ( -26.60) >DroAna_CAF1 31014 78 - 1 GCCGCGAGUCGUCAACAGGGAAACUUUCAGGAGGGCGCAGAUGUCAAAGACUUCAAGGACACGUUUCGUAGGUGGG--------UU----------------------------- ((((((((((.((...(((....)))....)).))).....((((...(....)...))))....)))).)))...--------..----------------------------- ( -18.90) >consensus GCUGCAAGCAGCCAGC___GCAAUGUACUUGGGGGAGCAGAAGCAAAAAACAAGAAUGAUCCAUUCCCUAGGUACGAAACAAGGUUUACC______UAAAG______________ ((((....))))...........((((((((((((.((....))......((....))......))))))))))))....................................... (-17.72 = -17.28 + -0.44)
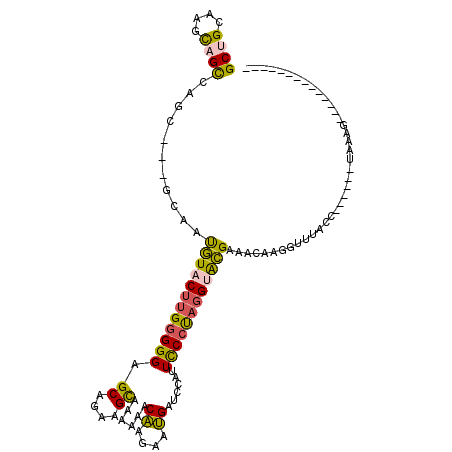
| Location | 148,333 – 148,445 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 83.72 |
| Mean single sequence MFE | -38.78 |
| Consensus MFE | -29.95 |
| Energy contribution | -30.15 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.623445 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 148333 112 - 22407834 CUCUGCCUCUCCCAAUACAAUUAAGAGGCGACGCAUGCGGGCUGCAAGCAGCCAGC---GCAAUGUACUUGGGGGAGCGGAAGCAAAAAACAAGAAUGAUCCAUUCCCCAGGUAC ...(((((((.............))))))).....(((((((((....)))))..)---)))..((((((((((((((....))......((....)).....)))))))))))) ( -44.42) >DroSec_CAF1 30777 112 - 1 CUCUGCCUCUCCCAAUACAAUUAAGAGGCGACGCAUGCGGGCUGCAAGCAGCCAGC---GCAAUGUACUUGGGGGAGCAGAAGCAAAAAACAAGAAUGAUCCAUUCCCUAGGUAC .(((((.((((((((((((.....(.(....).).(((((((((....)))))..)---))).)))).)))))))))))))............(((((...)))))......... ( -38.70) >DroSim_CAF1 32419 112 - 1 CUCUGCCUCUCCCAAUACAAUUAAGAGGCGACGCAUGCGGGCUGCAAGCAGCCAGC---GCAAUGUUCUUGGGGGAGCAGAAGCAAAAAACAAGAAUGAUCCAUUCCCUAGGUAC .(((((.((((((((.(((.....(.(....).).(((((((((....)))))..)---))).)))..)))))))))))))............(((((...)))))......... ( -36.90) >DroEre_CAF1 42411 112 - 1 CUCUGCCUCUCCCAAUACAAUUAAGAGGCGACGCAUGCGGGCGGCAACCAGCCAGC---GCGAUGUACUUGGGGGCGCGGAGGCUAAACGCAAGAAUGAUCCAUUCCCUAGGUAC ...(((((((.............))))))).(((..((.(((........))).))---)))..(((((((((((...(((..(....(....)...).)))..))))))))))) ( -38.12) >DroYak_CAF1 30973 112 - 1 CUCCGCCUCUCCCAACACAAUUAAGAGGCGUCGCAUGCGGGCUGCCAGCAGUCAGC---GCGAUGUACUUGGGGGCGCGGAAGCCAAGCACAAGAAUGAUCCAUUCCCUAGGUAC ....((((((.............))))))(((((..((.(((((....))))).))---)))))((((((((((((......)))........(((((...)))))))))))))) ( -42.82) >DroAna_CAF1 31017 115 - 1 CUCAGCCUCGCCGAACACCAUCAAGAGGCGUCGCAUGCGGGCCGCGAGUCGUCAACAGGGAAACUUUCAGGAGGGCGCAGAUGUCAAAGACUUCAAGGACACGUUUCGUAGGUGG ....(((((...((......))..)))))..(((.(((((..((.(.(((.((...(((....)))....)).)))...((.(((...))).)).....).))..))))).))). ( -31.70) >consensus CUCUGCCUCUCCCAAUACAAUUAAGAGGCGACGCAUGCGGGCUGCAAGCAGCCAGC___GCAAUGUACUUGGGGGAGCAGAAGCAAAAAACAAGAAUGAUCCAUUCCCUAGGUAC ....((((((.............))))))...((..((.(((((....))))).))...))...(((((((((((.((....))......((....))......))))))))))) (-29.95 = -30.15 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:23:48 2006