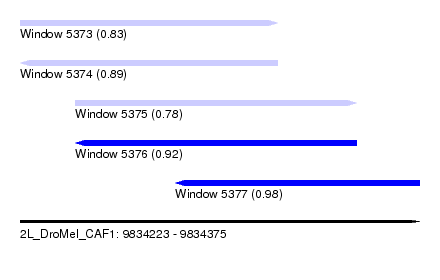
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,834,223 – 9,834,375 |
| Length | 152 |
| Max. P | 0.982233 |

| Location | 9,834,223 – 9,834,321 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -30.16 |
| Consensus MFE | -20.68 |
| Energy contribution | -20.20 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827675 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9834223 98 + 22407834 AAGUUUCUGAACCG-------CA--CCCCCUUUUUGGACCACAUCCAUGGGCAGUACGCCCAUGUG-GGAUGG-GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUA (((((.((....((-------(.--...((.....)).(((..(((((((((.....))))))).)-)..)))-)))((((((((......)))))))).)).))))). ( -34.70) >DroSec_CAF1 42774 82 + 1 AAGUUUCUAAACCA-------CU--CGCCCU--UUGGAC---------------UACGCCCAUGUGCGGGUGG-GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUA (((((.((...(((-------((--(((.(.--.(((..---------------.....))).).))))))))-...((((((((......)))))))).)).))))). ( -28.30) >DroSim_CAF1 45845 82 + 1 AAGUUUCUAAACCA-------CC--CGACCU--UUGGAC---------------UACGCCCAUGUGCGGGUGG-GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUA (((((.((...(((-------((--((....--.(((..---------------.....)))....)))))))-...((((((((......)))))))).)).))))). ( -28.60) >DroEre_CAF1 45316 95 + 1 AAGUUUCUGAACUU-------CUCCCCCCUU--UUGGACCGCAAGCA-GAGCACUACGCCCAUG----GGUGGGGCGGGACAAAUGGUGGAAUUUGUCCGAGUAAUUUA ..........((((-------(.(((((((.--.(((..(....)..-..((.....))))).)----)).)))).)((((((((......))))))))))))...... ( -28.30) >DroYak_CAF1 44832 97 + 1 AAGUUCCUGAACUUACCCAACCCCCCUCCCU--CUGGACCA----CA-GAGCACUACGCCCAUG----GGUGG-GCGGGGCAAAUGGUGAAAUUUGUCCGAGUAAUUUA (((((....)))))...........(((.((--(((.....----))-))).....((((((..----..)))-)))((((((((......)))))))))))....... ( -30.90) >consensus AAGUUUCUGAACCA_______CC__CCCCCU__UUGGACC_____CA_G_GCA_UACGCCCAUGUG_GGGUGG_GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUA (((((.((....................((.....))...................(((((((......)))).)))((((((((......)))))))).)).))))). (-20.68 = -20.20 + -0.48)


| Location | 9,834,223 – 9,834,321 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.16 |
| Mean single sequence MFE | -29.58 |
| Consensus MFE | -11.38 |
| Energy contribution | -11.14 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.38 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.886224 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9834223 98 - 22407834 UAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCAUCC-CACAUGGGCGUACUGCCCAUGGAUGUGGUCCAAAAAGGGGG--UG-------CGGUUCAGAAACUU ..........(((.....)))....((((..((((-((..((-(..((((((.....))))))((((...)))).....)))))--.)-------)))..))))..... ( -31.70) >DroSec_CAF1 42774 82 - 1 UAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCACCCGCACAUGGGCGUA---------------GUCCAA--AGGGCG--AG-------UGGUUUAGAAACUU ........(((((.....)))((((((((((((((-(((........))))))..---------------......--.)))))--))-------))))...))..... ( -28.80) >DroSim_CAF1 45845 82 - 1 UAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCACCCGCACAUGGGCGUA---------------GUCCAA--AGGUCG--GG-------UGGUUUAGAAACUU (((((((((((((.....)))(((.(((((..(((-(((........))))))..---------------)..)))--)))).)--))-------)))))))....... ( -25.60) >DroEre_CAF1 45316 95 - 1 UAAAUUACUCGGACAAAUUCCACCAUUUGUCCCGCCCCACC----CAUGGGCGUAGUGCUC-UGCUUGCGGUCCAA--AAGGGGGGAG-------AAGUUCAGAAACUU ..........((((...((((.((.((((..((((((((..----..)))).((((....)-)))..))))..)))--).)).)))).-------..))))........ ( -30.40) >DroYak_CAF1 44832 97 - 1 UAAAUUACUCGGACAAAUUUCACCAUUUGCCCCGC-CCACC----CAUGGGCGUAGUGCUC-UG----UGGUCCAG--AGGGAGGGGGGUUGGGUAAGUUCAGGAACUU .................((((...((((((((.((-((.((----(..((((.....))))-..----...(((..--..)))))))))).))))))))...))))... ( -31.40) >consensus UAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC_CCACCC_CACAUGGGCGUA_UGC_C_UG_____GGUCCAA__AGGGCG__AG_______AGGUUCAGAAACUU ..........((((((((......))))....(((.(((........)))))).................))))......................((((....)))). (-11.38 = -11.14 + -0.24)


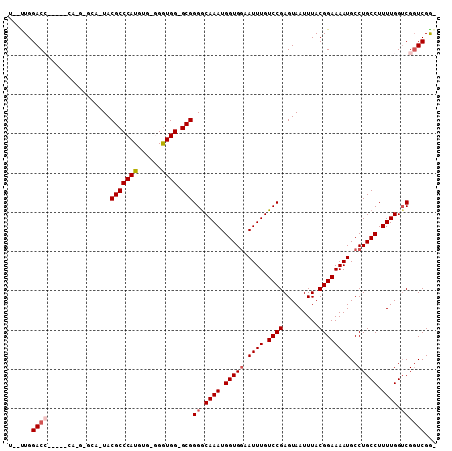
| Location | 9,834,244 – 9,834,351 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -25.18 |
| Energy contribution | -26.02 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784875 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9834244 107 + 22407834 UUUUUGGACCACAUCCAUGGGCAGUACGCCCAUGUG-GGAUGG-GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUACGGAAAAUGCCUGCCUUUUGGUCGUUCGAG ......((((((((((((((((.....))))))...-)))))(-(((((.(((((......)))))((((.........)))).....))))))...)))))....... ( -40.10) >DroSec_CAF1 42795 91 + 1 U--UUGGAC---------------UACGCCCAUGUGCGGGUGG-GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUACGGAAAAUGCCUGCCUUUUGGUCGGUCGGA .--((.(((---------------(.(((((((......))))-)))((.((((.((..(.((((.((((.........))))))))..)..)).)))).)))))).)) ( -33.30) >DroSim_CAF1 45866 91 + 1 U--UUGGAC---------------UACGCCCAUGUGCGGGUGG-GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUACGGAAAAUGCCUGCCUUUUGGUCGGUCGGA .--((.(((---------------(.(((((((......))))-)))((.((((.((..(.((((.((((.........))))))))..)..)).)))).)))))).)) ( -33.30) >DroEre_CAF1 45339 101 + 1 U--UUGGACCGCAAGCA-GAGCACUACGCCCAUG----GGUGGGGCGGGACAAAUGGUGGAAUUUGUCCGAGUAAUUUACGGAAAAUGCCUGCCUUUUGGUCGGUCGG- .--...(((((.(((((-(.(((...(((((...----....)))))((((((((......)))))))).................)))))).))).)))))......- ( -37.10) >DroYak_CAF1 44862 96 + 1 U--CUGGACCA----CA-GAGCACUACGCCCAUG----GGUGG-GCGGGGCAAAUGGUGAAAUUUGUCCGAGUAAUUUACGGAAAAUGCCUGCCUUUUGGACGGUCGG- .--...((((.----((-(.(((...((((((..----..)))-)))((((((((......)))))))).................))))))((....))..))))..- ( -35.10) >consensus U__UUGGACC_____CA_G_GCA_UACGCCCAUGUG_GGGUGG_GCGGGGCAAAUGGUGGAAUUUGUCCGAGUAAUUUACGGAAAAUGCCUGCCUUUUGGUCGGUCGG_ ......((((................(((((((......)))).)))((.((((.(((((.((((.((((.........))))))))..))))).)))).))))))... (-25.18 = -26.02 + 0.84)

| Location | 9,834,244 – 9,834,351 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 83.80 |
| Mean single sequence MFE | -28.46 |
| Consensus MFE | -18.24 |
| Energy contribution | -19.00 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.919654 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9834244 107 - 22407834 CUCGAACGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCAUCC-CACAUGGGCGUACUGCCCAUGGAUGUGGUCCAAAAA .......(((((...(((.((((......(((...)))..(((.....)))......))))..))-)(((((-...((((((.....))))))))))))))))...... ( -33.40) >DroSec_CAF1 42795 91 - 1 UCCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCACCCGCACAUGGGCGUA---------------GUCCAA--A .......(((.....((((((.((((((((.........)))).)))).))......)))).(((-(((........))))))..---------------)))...--. ( -22.30) >DroSim_CAF1 45866 91 - 1 UCCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCACCCGCACAUGGGCGUA---------------GUCCAA--A .......(((.....((((((.((((((((.........)))).)))).))......)))).(((-(((........))))))..---------------)))...--. ( -22.30) >DroEre_CAF1 45339 101 - 1 -CCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGUCCCGCCCCACC----CAUGGGCGUAGUGCUC-UGCUUGCGGUCCAA--A -..(((((......(((((((((((...............((((((((......))))))))(((((....----...))))).))))).)-))))).)))))...--. ( -37.60) >DroYak_CAF1 44862 96 - 1 -CCGACCGUCCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUUCACCAUUUGCCCCGC-CCACC----CAUGGGCGUAGUGCUC-UG----UGGUCCAG--A -..((((...(....)(((((((((...............((.(((((......))))))).(((-(((..----..)))))).))))).)-))----)))))...--. ( -26.70) >consensus _CCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC_CCACCC_CACAUGGGCGUA_UGC_C_UG_____GGUCCAA__A ...(((((..((((.((..((.((((((((.........)))).)))).)).)).))))...(((.(((........))))))...............)))))...... (-18.24 = -19.00 + 0.76)



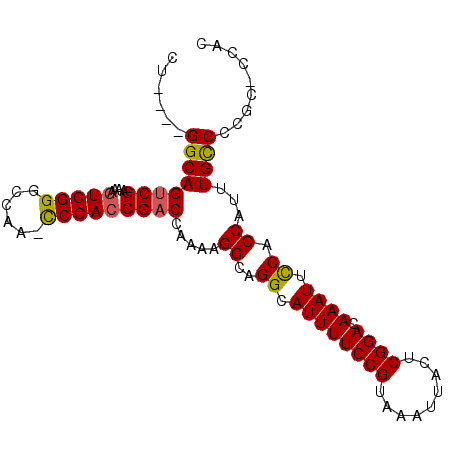
| Location | 9,834,282 – 9,834,375 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 93.76 |
| Mean single sequence MFE | -24.92 |
| Consensus MFE | -21.60 |
| Energy contribution | -21.52 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9834282 93 - 22407834 CU----GGCAGUCGAAAAGUCGGGCCAACUCGAACGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCAU ..----(((.((((.....(((((....))))).)))).....((((((.((((((((.........)))).)))).))......))))..))-)... ( -23.50) >DroSec_CAF1 42817 93 - 1 CU----GGCAGUCGAAAAGUCGGGCCAAUCCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCAC ..----(((.((((....((((((....)))))))))).....((((((.((((((((.........)))).)))).))......))))..))-)... ( -25.90) >DroSim_CAF1 45888 93 - 1 CU----GGCAGUCGAAAAGUCGGGCCAAUCCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC-CCAC ..----(((.((((....((((((....)))))))))).....((((((.((((((((.........)))).)))).))......))))..))-)... ( -25.90) >DroEre_CAF1 45371 97 - 1 CUAACAGGCAGUCGAAAAGUCGGGCCAA-CCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGUCCCGCCCCAC ......(((.((((....(((((.....-)))))))))((((.((..((.((((((((.........)))).)))).)).)).))))....))).... ( -26.90) >DroYak_CAF1 44890 96 - 1 CUAACAGGCAGUCGAAAAGUCGGGCCAA-CCGACCGUCCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUUCACCAUUUGCCCCGC-CCAC ......(((....((...(((((.....-)))))..)).....((((((.((((((((.........)))).))))....))...))))..))-)... ( -22.40) >consensus CU____GGCAGUCGAAAAGUCGGGCCAA_CCGACCGACCAAAAGGCAGGCAUUUUCCGUAAAUUACUCGGACAAAUUCCACCAUUUGCCCCGC_CCAC ......((((((((....(((((......))))))))).....((..((.((((((((.........)))).)))).)).))...))))......... (-21.60 = -21.52 + -0.08)

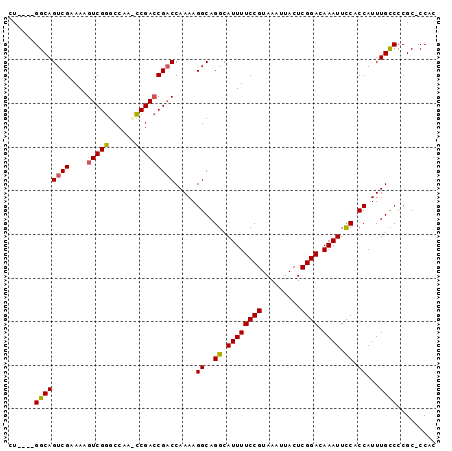
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:30 2006