| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,829,119 – 9,829,226 |
| Length | 107 |
| Max. P | 0.866063 |

| Location | 9,829,119 – 9,829,226 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -20.95 |
| Energy contribution | -20.40 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.866063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
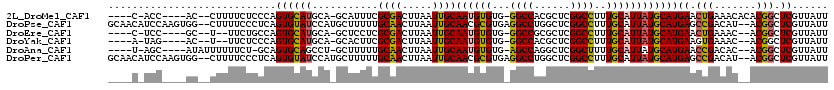
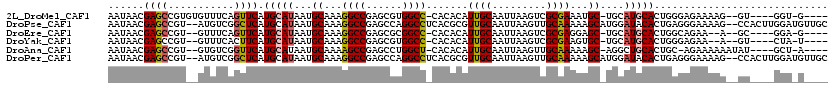
>2L_DroMel_CAF1 9829119 107 + 22407834 ----C-ACC----AC--CUUUUCUCCCAGUGCAUGCA-GCAUUUCGCGACUUAAUUGCAAUGUGUG-GGCCACGCUCGGCCUUUGCAUUAUGCAUGAACUGAAACACACGGCUCGUUAUU ----.-...----..--.........((((.((((((-.......((((.....))))((((((.(-((((......))))).)))))).)))))).))))................... ( -29.40) >DroPse_CAF1 39647 116 + 1 GCAACAUCCAAGUGG--CUUUUCCCUCAGUGUAUCCAUGCUUUUUGCAACUUAAUUGCAACGCGUGAGGCCUGGCUCGGCCUUUGCAUUAUGCAUGAGCCGACAU--ACGGCUCGUUAUU (((........(.((--......)).)((((((..(((((...((((((.....)))))).)))))(((((......))))).)))))).)))((((((((....--.)))))))).... ( -37.90) >DroEre_CAF1 40161 103 + 1 ----C-UCC----GC--U--UUCUGCCAGUGCAUGCA-GCUCCUCGCGACUUAAUUGCAAUGUGUG-GGCCGCGCUCGGCCUUUGCAUUAUGCAUGAACUGAAAC--ACGGCUCGUUAUU ----.-...----((--(--......((((.((((((-.......((((.....))))((((((.(-(((((....)))))).)))))).)))))).))))....--..)))........ ( -32.80) >DroYak_CAF1 38601 103 + 1 ----A-UAG----AC--U--UUCUCCCAGUGCAUGCA-GCACUUCGCGACUUAAUUGCAAUGUGUG-GGCCACGCUCGGCCUUUGCAUUAUGCAUGAAGUGAAAC--ACGGCUCGUUAUU ----.-...----..--.--........(.((.....-.((((((((((.....))))((((((.(-((((......))))).))))))......))))))....--...)).)...... ( -27.22) >DroAna_CAF1 38190 106 + 1 ----U-AGC----AUAUUUUUUCU-GCAGUGCAGCCU-GCUUUUUGCAACUUAAUUGCAAUGUGUG-AGCCAGGCUCGGCUUUUGCAUUAUGCAUGAACCGACAC--ACGGCUCGUUAUU ----.-.((----(((.......(-((((.(((((((-((((.((((((.....)))))).....)-)).))))))..))..))))).)))))((((.(((....--.))).)))).... ( -31.20) >DroPer_CAF1 40225 116 + 1 GCAACAUCCAAGUGG--CUUUUCCCUCAGUGUAUCCAUGCUUUUUGCAACUUAAUUGCAACGCGUGAGGCCUGGCUCGGCCUUUGCAUUAUGCAUGAGCCGACAU--ACGGCUCGUUAUU (((........(.((--......)).)((((((..(((((...((((((.....)))))).)))))(((((......))))).)))))).)))((((((((....--.)))))))).... ( -37.90) >consensus ____C_UCC____AC__CUUUUCUCCCAGUGCAUCCA_GCUUUUCGCAACUUAAUUGCAAUGUGUG_GGCCACGCUCGGCCUUUGCAUUAUGCAUGAACCGAAAC__ACGGCUCGUUAUU ............................((((((...........((((.....))))((((((...((((......))))..))))))))))))((.(((.......))).))...... (-20.95 = -20.40 + -0.55)
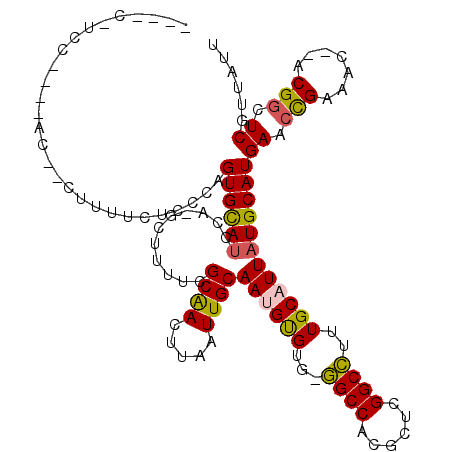
| Location | 9,829,119 – 9,829,226 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -34.20 |
| Consensus MFE | -16.32 |
| Energy contribution | -16.23 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535244 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
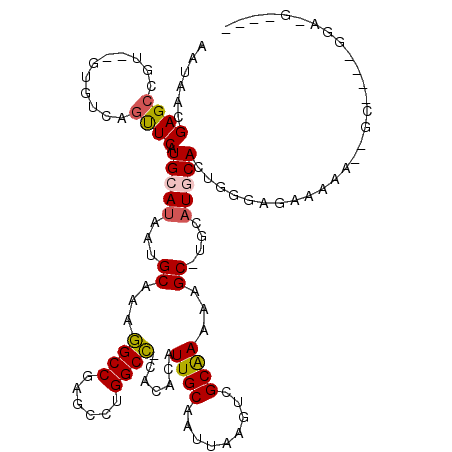
>2L_DroMel_CAF1 9829119 107 - 22407834 AAUAACGAGCCGUGUGUUUCAGUUCAUGCAUAAUGCAAAGGCCGAGCGUGGCC-CACACAUUGCAAUUAAGUCGCGAAAUGC-UGCAUGCACUGGGAGAAAAG--GU----GGU-G---- ....((..(((...(.(..((((.((((((....(((..(((((....)))))-......((((.........))))..)))-)))))).))))..).)...)--))----.))-.---- ( -32.70) >DroPse_CAF1 39647 116 - 1 AAUAACGAGCCGU--AUGUCGGCUCAUGCAUAAUGCAAAGGCCGAGCCAGGCCUCACGCGUUGCAAUUAAGUUGCAAAAAGCAUGGAUACACUGAGGGAAAAG--CCACUUGGAUGUUGC ..((((((((((.--....))))))((((..(((((..(((((......)))))...)))))(((((...))))).....)))).......(..((((.....--)).))..)..)))). ( -39.40) >DroEre_CAF1 40161 103 - 1 AAUAACGAGCCGU--GUUUCAGUUCAUGCAUAAUGCAAAGGCCGAGCGCGGCC-CACACAUUGCAAUUAAGUCGCGAGGAGC-UGCAUGCACUGGCAGAA--A--GC----GGA-G---- ........(((((--((..((((((.((((..(((....(((((....)))))-....))))))).......(....)))))-))...)))).)))....--.--..----...-.---- ( -32.40) >DroYak_CAF1 38601 103 - 1 AAUAACGAGCCGU--GUUUCACUUCAUGCAUAAUGCAAAGGCCGAGCGUGGCC-CACACAUUGCAAUUAAGUCGCGAAGUGC-UGCAUGCACUGGGAGAA--A--GU----CUA-U---- .....(.((.(((--((..((((((.(((.((((((((.(((((....)))))-......)))).)))).).)).)))))).-.)))))..)).).....--.--..----...-.---- ( -26.20) >DroAna_CAF1 38190 106 - 1 AAUAACGAGCCGU--GUGUCGGUUCAUGCAUAAUGCAAAAGCCGAGCCUGGCU-CACACAUUGCAAUUAAGUUGCAAAAAGC-AGGCUGCACUGC-AGAAAAAAUAU----GCU-A---- ......((((((.--((.((((((..(((.....)))..)))))))).)))))-).....(((((((...)))))))...((-((......))))-...........----...-.---- ( -35.10) >DroPer_CAF1 40225 116 - 1 AAUAACGAGCCGU--AUGUCGGCUCAUGCAUAAUGCAAAGGCCGAGCCAGGCCUCACGCGUUGCAAUUAAGUUGCAAAAAGCAUGGAUACACUGAGGGAAAAG--CCACUUGGAUGUUGC ..((((((((((.--....))))))((((..(((((..(((((......)))))...)))))(((((...))))).....)))).......(..((((.....--)).))..)..)))). ( -39.40) >consensus AAUAACGAGCCGU__GUGUCAGUUCAUGCAUAAUGCAAAGGCCGAGCCUGGCC_CACACAUUGCAAUUAAGUCGCAAAAAGC_UGCAUGCACUGGGAGAAAAA__GC____GGA_G____ ......((((...........)))).(((((...((...((((......)))).......((((.........))))...))....)))))............................. (-16.32 = -16.23 + -0.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:24 2006