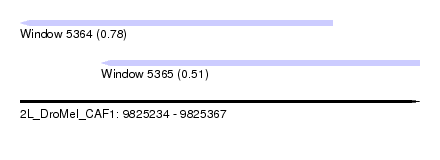
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,825,234 – 9,825,367 |
| Length | 133 |
| Max. P | 0.782193 |

| Location | 9,825,234 – 9,825,338 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 76.44 |
| Mean single sequence MFE | -23.55 |
| Consensus MFE | -15.97 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782193 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
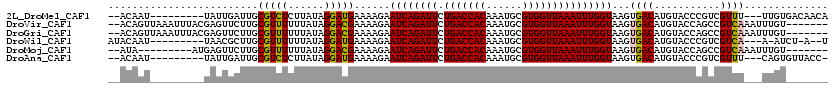
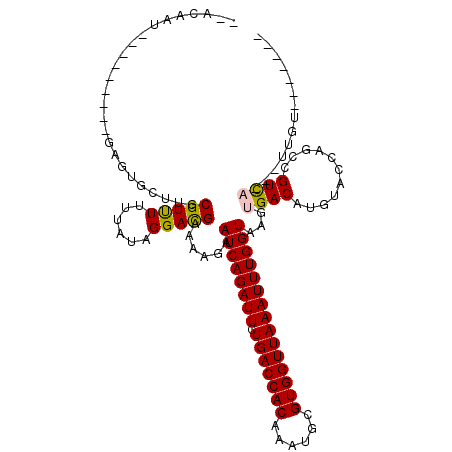
>2L_DroMel_CAF1 9825234 104 - 22407834 GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU---UUGUGACAACACAAUAUCACAACUAUU---CUUUUGUGAUU------ .((((((((((((((.(((((((......)))))))))))))))..((((..(((....((((...---...))))...)))...))))......)---)))))......------ ( -25.20) >DroVir_CAF1 46757 102 - 1 GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCAGCCGUCAAAUUUGU-----------CUAGCACCUGUU---UGUCUGUCUUUCUUAUC ((.(((((.(((((..(((((((......))))))).....(((....((((..................)))-----------)....)))....---.)))))...))))).)) ( -21.87) >DroPse_CAF1 36077 104 - 1 GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU---GUGUGUC---UCAUCUUCACAGCUUUCUUGCUUCUUUCAUU------ ((((.(((..((((...(((((((((((((.(((......((........))...))))).)))))---))).)))---..)))).....((......)))))))))...------ ( -24.80) >DroWil_CAF1 49323 100 - 1 GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUCA---A-AUCU-A--UC-AUUUAGCACCUGUU---GUGUGUCAAUU---U-- .......((((((((.(((((((......)))))))))))))))...(((((..((.(.(..(..(---(-((..-.--..-))))..)..).).)---)..)))))...---.-- ( -23.50) >DroAna_CAF1 33925 98 - 1 GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU---CAGUGUUACC-----AUCACAACUAAU---UA-UUAGGCUU------ .....((((((((((.(((((((......)))))))))))))))..(((((((..((.....))..---..))))))).-----.)).........---..-........------ ( -21.10) >DroPer_CAF1 36599 104 - 1 GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU---GUGUGUC---UCAUCUUCACAGCUUUCUUGCUUCUUUCAUU------ ((((.(((..((((...(((((((((((((.(((......((........))...))))).)))))---))).)))---..)))).....((......)))))))))...------ ( -24.80) >consensus GAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU___GUGUGUC___UC___UUCACAACUAUU___CUUCUUUCAUU______ .......((((((((.(((((((......)))))))))))))))....(((........)))...................................................... (-15.97 = -16.13 + 0.17)

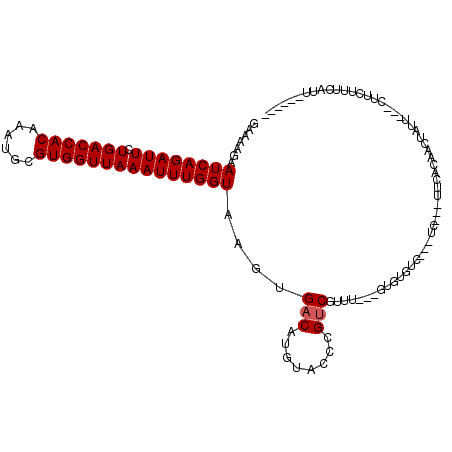
| Location | 9,825,261 – 9,825,367 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -24.13 |
| Consensus MFE | -18.39 |
| Energy contribution | -18.03 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.76 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.512144 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9825261 106 - 22407834 --ACAAU---------UAUUGAUUGCGUCUCUUAUAGGAUGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU---UUGUGACAACA --.((((---------(...)))))(((((......)))))......((((((((.(((((((......)))))))))))))))..((.(((...((.....))..---.))).)).... ( -22.50) >DroVir_CAF1 46786 111 - 1 --ACAGUUAAAUUUACGAGUUCUUGCGUUUUUUAUAGGACGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCAGCCGUCAAAUUUGU------- --((((((..((((((..((((((.((((((....))))))..))))))((((((.(((((((......)))))))))))))))))))))).)))..................------- ( -26.00) >DroGri_CAF1 37820 111 - 1 --ACAGUUAAAUUUACGAGUUCUUGCGUUUUUUAUAGGACGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCAGCCGUCAAAUUUGU------- --((((((..((((((..((((((.((((((....))))))..))))))((((((.(((((((......)))))))))))))))))))))).)))..................------- ( -26.00) >DroWil_CAF1 49350 104 - 1 AUACAAU---------UAACGCUUGCGUUUUUUAUAGGAUGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUCA---A-AUCU-A--U ....((.---------.((((....))))..))...((((.......((((((((.(((((((......)))))))))))))))...(((((((....))).))))---.-))))-.--. ( -23.30) >DroMoj_CAF1 43821 102 - 1 --AUA---------AUGAGUUCUUGCGUUUUUUAUAGGACGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCAGCCGUCAAAUUUGU------- --...---------......((((.((((((....))))))..))))((((((((.(((((((......)))))))))))))))...((((..((....)).)))).......------- ( -22.70) >DroAna_CAF1 33947 105 - 1 --ACAAU---------UAUUGAUUGCGUCUCUUAUAGGAUGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCCGUCGUUU---CAGUGUUACC- --.((((---------(...)))))(((((......)))))......((((((((.(((((((......)))))))))))))))..(((((((..((.....))..---..))))))).- ( -24.30) >consensus __ACAAU_________GAGUGCUUGCGUUUUUUAUAGGACGAAAAGAAUCAGAUUCUGACCACAAAUGCGUGGUUAAAUUUGGUAAGUGACAUGUACCAGCCGUCA___UUGU_______ .........................(((((......)))))......((((((((.(((((((......)))))))))))))))...((((...........)))).............. (-18.39 = -18.03 + -0.36)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:18 2006