
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,817,760 – 9,817,867 |
| Length | 107 |
| Max. P | 0.567304 |

| Location | 9,817,760 – 9,817,867 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 81.00 |
| Mean single sequence MFE | -27.37 |
| Consensus MFE | -16.55 |
| Energy contribution | -16.69 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9817760 107 - 22407834 AUGCCAGUCAACCGAUUGUUGUUCUGCUCGACGGCCCAAUUAUGCCGUCAAAUAGCAAUUAAAAGCGGCAAAAUGAACG-CAAAGGCCGAAAGGAACAAAAGCGC-GAC------ .((((((((....)))))(((((((..((((((((........)))))).....((........))(((....((....-))...)))))..)))))))....))-)..------ ( -30.40) >DroPse_CAF1 28144 111 - 1 GUGUCCGUCAGCCAAAUUUUGUUCUGCUCGACGGCUCAAUUAUGCCGUGUAUUGGCAAUUAAAACCGCUA-AACGAACG-CCAAAACAAUCACAACUGA-AGUGC-GGAAUGAUA ..................(..((((((.(.(((((........)))))((.(((((..............-.......)-)))).))............-.).))-))))..).. ( -25.00) >DroSim_CAF1 29598 106 - 1 AUGCCAGUCAACCGAUUGUUGUUCUGCUCGACGGCCCAAUUAUGCCGUCAAAUAGCAAUUAAAAGCGGCA-AAUGAACG-CAAAGGCCAACAGGAACAAAAGCGC-GAC------ .((((((((....)))))((((((((((.((((((........))))))....)))..........(((.-..((....-))...)))....)))))))....))-)..------ ( -29.20) >DroEre_CAF1 29044 105 - 1 AUGCCAGUCAACCGAUUGUUGUUCUGCUCGACGGCCCAAUUAUGCCGUCAAAUAGCAAUUAAAAGCGGCA-AAUGAACG-CAAAGGCCAUCUCGAACAA-AGCGC-GAC------ .((((((((....)))))((((((((((.((((((........))))))....)))).........(((.-..((....-))...))).....))))))-...))-)..------ ( -28.90) >DroYak_CAF1 27010 105 - 1 AUGCCAGUCAACCGAUUGUUGUUCUGCUCGACGGCCCAAUUAUGCCGUCAAAUGGCAAUUAAAAGCGGCA-AAUGAACG-CAAAGGCCAACGCGAACAA-AGCGC-GAC------ .((((((((....)))))((((((.((..((((((........))))))...((((........(((...-......))-)....))))..))))))))-...))-)..------ ( -31.60) >DroAna_CAF1 26816 99 - 1 ---------AGCCGAUUGUAGUUCUGCUCGACGGCCCAAUUAUGCCGUCAAAUAGCAAUUAAAACCGAUA-AAUGAACACCAAAAACCAUCUGGACCAAAAACGCAGAC------ ---------.............(((((..((((((........)))))).....................-...............((....)).........))))).------ ( -19.10) >consensus AUGCCAGUCAACCGAUUGUUGUUCUGCUCGACGGCCCAAUUAUGCCGUCAAAUAGCAAUUAAAAGCGGCA_AAUGAACG_CAAAGGCCAACACGAACAA_AGCGC_GAC______ ............((....((((((((((.((((((........))))))....)))).........(((................))).....))))))...))........... (-16.55 = -16.69 + 0.14)
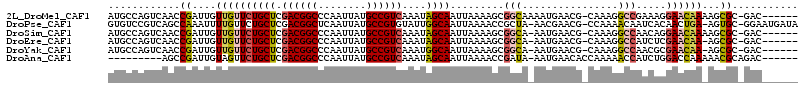
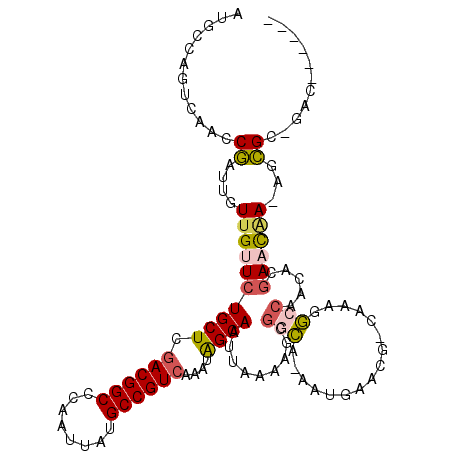

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:16 2006