| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,815,316 – 9,815,406 |
| Length | 90 |
| Max. P | 0.997257 |

| Location | 9,815,316 – 9,815,406 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 73.57 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -17.24 |
| Energy contribution | -18.44 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -4.23 |
| Structure conservation index | 0.60 |
| SVM decision value | 2.83 |
| SVM RNA-class probability | 0.997257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
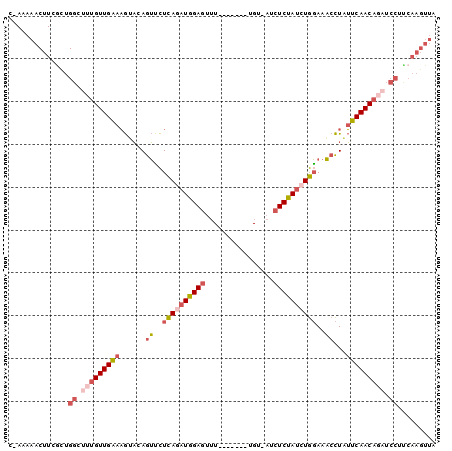
>2L_DroMel_CAF1 9815316 90 + 22407834 C-AAAAACUUCGUUGGCUUUGUUGAAAGUACAGUUCUCAGAUGGAGU-U-------UGU-AUCUCUAUCUGGAAACCUAUUCAACAGAUCCUUCAAGUUA .-...(((((.(..((.(((((((((((....(((..(((((((((.-.-------...-..)))))))))..))))).))))))))).))..)))))). ( -29.70) >DroSec_CAF1 23986 91 + 1 C-AAAAACUUCGCUGGCUUUGUUGAAAGUACAGUCCUCAGAUGGAGUUU-------UGU-AUCUCUAUCUGGAAACCUAUUCAACAGAUCCUUCAAGUUA .-...(((((.(..((.(((((((((((....((..((((((((((...-------...-..))))))))))..)))).))))))))).))..)))))). ( -27.60) >DroSim_CAF1 27192 91 + 1 C-AAAAACUUCGCUGGCUUUGUUGAAAGUACAGUCCUCAGAUGGAGUUU-------UGU-AUCUCUAUCUGGAAACCUAUUCAACAGAUCCUUCAAGAUA .-.....(((.(..((.(((((((((((....((..((((((((((...-------...-..))))))))))..)))).))))))))).))..))))... ( -25.60) >DroEre_CAF1 26679 96 + 1 C-AAAAACUUAGCUGGGUUUGUUGAAAGCACAGUUCUCAGAUGGAGUUUGUACU--CGU-AUCUCUAUCUGGAAACCUAUUCAACAGAUCCGCCAAGUUA .-...(((((.((.((((((((((((((....(((..(((((((((..((....--)).-..)))))))))..))))).)))))))))))))).))))). ( -37.00) >DroYak_CAF1 24549 90 + 1 --AAAAACUUGACUGGGCGAGUUGCAAGUACAGUUCUCAGAUGGAGUUU-------UGU-AGCUCUAUGUGGAAACCUAUGCAACAGAUCCUUCAAGUGA --....((((((..((((..((((((((....(((..((.((((((((.-------...-)))))))).))..))))).)))))).).))).)))))).. ( -30.60) >DroAna_CAF1 24509 89 + 1 CAAUAAACGUAAUCGAGAUUGUUGCAUAAUCAAGAUUUAAAUGGAACUUACAGUUAAGUAAGCUCCAAAUAUAAUCUUUGGCAACA-----------UUC ................((.((((((......((((((....((((.(((((......))))).)))).....))))))..))))))-----------.)) ( -21.30) >consensus C_AAAAACUUCGCUGGCUUUGUUGAAAGUACAGUUCUCAGAUGGAGUUU_______UGU_AUCUCUAUCUGGAAACCUAUUCAACAGAUCCUUCAAGUUA ..............((.(((((((((......((..((((((((((................))))))))))..))...))))))))).))......... (-17.24 = -18.44 + 1.20)


| Location | 9,815,316 – 9,815,406 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 73.57 |
| Mean single sequence MFE | -26.37 |
| Consensus MFE | -17.06 |
| Energy contribution | -17.26 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -3.40 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.997033 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
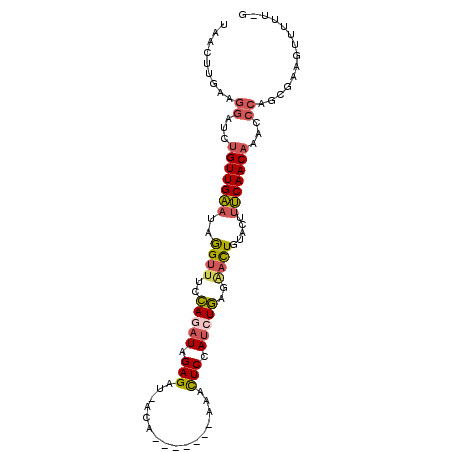
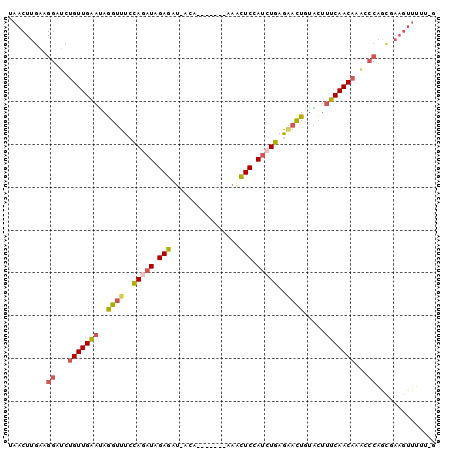
>2L_DroMel_CAF1 9815316 90 - 22407834 UAACUUGAAGGAUCUGUUGAAUAGGUUUCCAGAUAGAGAU-ACA-------A-ACUCCAUCUGAGAACUGUACUUUCAACAAAGCCAACGAAGUUUUU-G .((((((..((...(((((((..((((..(((((.(((..-...-------.-.))).)))))..)))).....)))))))...))..).)))))...-. ( -25.20) >DroSec_CAF1 23986 91 - 1 UAACUUGAAGGAUCUGUUGAAUAGGUUUCCAGAUAGAGAU-ACA-------AAACUCCAUCUGAGGACUGUACUUUCAACAAAGCCAGCGAAGUUUUU-G .((((((..((...(((((((..((((..(((((.(((..-...-------...))).)))))..)))).....)))))))...))..).)))))...-. ( -26.00) >DroSim_CAF1 27192 91 - 1 UAUCUUGAAGGAUCUGUUGAAUAGGUUUCCAGAUAGAGAU-ACA-------AAACUCCAUCUGAGGACUGUACUUUCAACAAAGCCAGCGAAGUUUUU-G ....(((..((...(((((((..((((..(((((.(((..-...-------...))).)))))..)))).....)))))))...))..))).......-. ( -24.90) >DroEre_CAF1 26679 96 - 1 UAACUUGGCGGAUCUGUUGAAUAGGUUUCCAGAUAGAGAU-ACG--AGUACAAACUCCAUCUGAGAACUGUGCUUUCAACAAACCCAGCUAAGUUUUU-G .((((((((((...(((((((..((((..(((((.(((..-...--........))).)))))..)))).....)))))))...)).))))))))...-. ( -30.52) >DroYak_CAF1 24549 90 - 1 UCACUUGAAGGAUCUGUUGCAUAGGUUUCCACAUAGAGCU-ACA-------AAACUCCAUCUGAGAACUGUACUUGCAACUCGCCCAGUCAAGUUUUU-- ..((((((.((....((((((..((((..((.((.(((..-...-------...))).)).))..)))).....))))))...))...))))))....-- ( -23.70) >DroAna_CAF1 24509 89 - 1 GAA-----------UGUUGCCAAAGAUUAUAUUUGGAGCUUACUUAACUGUAAGUUCCAUUUAAAUCUUGAUUAUGCAACAAUCUCGAUUACGUUUAUUG ((.-----------((((((..((((((.((..((((((((((......))))))))))..))))))))......)))))).))................ ( -27.90) >consensus UAACUUGAAGGAUCUGUUGAAUAGGUUUCCAGAUAGAGAU_ACA_______AAACUCCAUCUGAGAACUGUACUUUCAACAAACCCAGCGAAGUUUUU_G .........((...(((((((..((((..(((((.(((................))).)))))..)))).....)))))))...)).............. (-17.06 = -17.26 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:14 2006