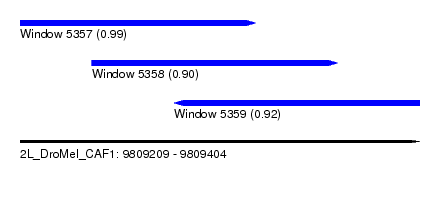
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,809,209 – 9,809,404 |
| Length | 195 |
| Max. P | 0.994154 |

| Location | 9,809,209 – 9,809,324 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -25.43 |
| Consensus MFE | -25.10 |
| Energy contribution | -25.10 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.99 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9809209 115 + 22407834 UUGC-----CACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA ....-----.((.((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).)))).)).((((((.((........))))))))... ( -25.10) >DroVir_CAF1 26951 114 + 1 -AAA-----CACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA -...-----.((.((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).)))).)).((((((.((........))))))))... ( -25.10) >DroGri_CAF1 20059 114 + 1 -AAA-----CACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA -...-----.((.((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).)))).)).((((((.((........))))))))... ( -25.10) >DroWil_CAF1 25913 120 + 1 UUGCCAAAACACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA ..........((.((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).)))).)).((((((.((........))))))))... ( -25.10) >DroMoj_CAF1 21557 114 + 1 -AAA-----CACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA -...-----.((.((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).)))).)).((((((.((........))))))))... ( -25.10) >DroAna_CAF1 18369 120 + 1 UUGCUUUUGCACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA .(((....)))..((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).))))(((((.((..((....))....)))))))... ( -27.10) >consensus _AAA_____CACUUUACCUGAUUUCCAUCGUAAGUCCAACUACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCA ..........((.((((.((.(((((((.....(.(((((...((((......))))....))))).).....))))))).)).)))).)).((((((.((........))))))))... (-25.10 = -25.10 + -0.00)

| Location | 9,809,244 – 9,809,364 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -32.20 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9809244 120 + 22407834 UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... ( -32.20) >DroWil_CAF1 25953 120 + 1 UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... ( -32.20) >DroYak_CAF1 18445 120 + 1 UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... ( -32.20) >DroMoj_CAF1 21591 120 + 1 UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... ( -32.20) >DroAna_CAF1 18409 120 + 1 UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... ( -32.20) >DroPer_CAF1 19708 120 + 1 UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... ( -32.20) >consensus UACCGAAUUUCAUUUCGCAAUGUUGGUCAUCAAAUGGGAACCACGUGAUGUCUAUCUUGCAUGUGCUCUUGAAGAUACCAGGGGGCACGUACAGACAACUGCCGUUAUGUUUGACCACAA ....(((......)))((((.(.(((.(((((..(((...)))..))))).))).)))))((((((((((..........)))))))))).((((((((....))..))))))....... (-32.20 = -32.20 + -0.00)
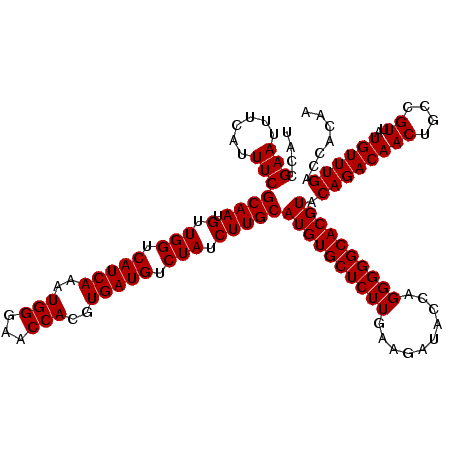
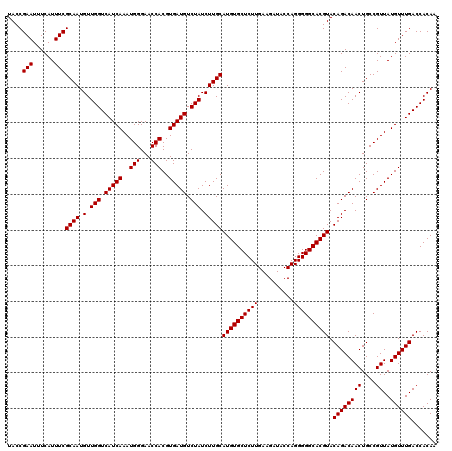
| Location | 9,809,284 – 9,809,404 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 100.00 |
| Mean single sequence MFE | -37.01 |
| Consensus MFE | -37.01 |
| Energy contribution | -37.01 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.97 |
| Structure conservation index | 1.00 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9809284 120 - 22407834 AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). ( -37.01) >DroWil_CAF1 25993 120 - 1 AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). ( -37.01) >DroYak_CAF1 18485 120 - 1 AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). ( -37.01) >DroMoj_CAF1 21631 120 - 1 AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). ( -37.01) >DroAna_CAF1 18449 120 - 1 AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). ( -37.01) >DroPer_CAF1 19748 120 - 1 AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). ( -37.01) >consensus AGCGCGGAUGAGGGAUUCGAUGGCACGUAUCACACCAACAUUGUGGUCAAACAUAACGGCAGUUGUCUGUACGUGCCCCCUGGUAUCUUCAAGAGCACAUGCAAGAUAGACAUCACGUGG ..((((((((((((.......((((((((.((...((((..(((.((........)).)))))))..))))))))))))))..((((((((........)).))))))..)))).)))). (-37.01 = -37.01 + 0.00)

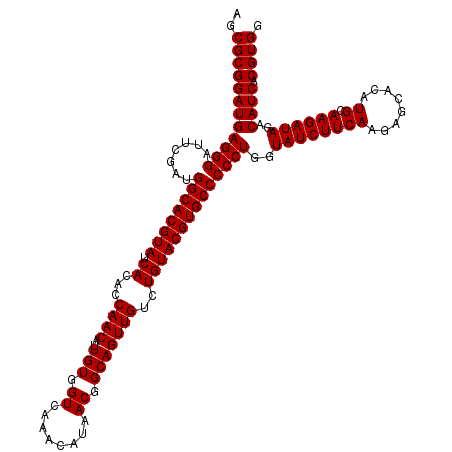
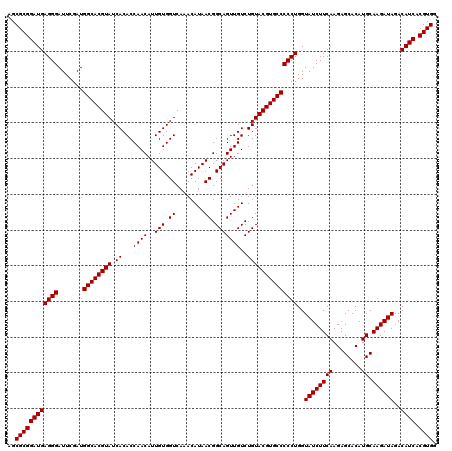
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:12 2006