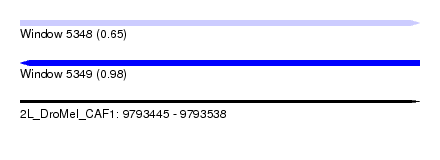
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,793,445 – 9,793,538 |
| Length | 93 |
| Max. P | 0.975460 |

| Location | 9,793,445 – 9,793,538 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -15.82 |
| Consensus MFE | -11.70 |
| Energy contribution | -11.70 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650830 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
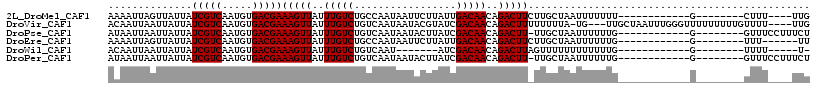
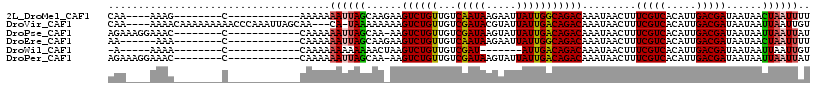
>2L_DroMel_CAF1 9793445 93 + 22407834 AAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUUGACAACAGACUUCUUGCUAAUUUUUUU------------G--------CUUU----UUG (((((((((.....(((((.....))))).........(((((.(((((.....)))))....))))).....)))))))))...------------.--------....----... ( -18.40) >DroVir_CAF1 2730 109 + 1 ACAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACGUAUCGACAACAGACUUUUUUUUA-UG---UUGCUAAUUUGGGUUUUUUUUUGUUUU----UUG ((((..((..(((.....((((((((.(((((((...(((.((((.(((.....))).))))))))))))))..)))-))---)))........)))..))..))))...----... ( -14.82) >DroPse_CAF1 2785 96 + 1 AUAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACUUAUCGACAACAGACUU-UUGCUAAUUUUUUG------------G--------GUUUCCUUUCU .(((..(((((.((..(((...(((((....))))).(((.((((.(((.....))).))))))))))..-.)).)))))..)))------------.--------........... ( -14.70) >DroEre_CAF1 5040 91 + 1 AAAAUUAGUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGCCAAUAAUUCUUAUUGACAACAGACUUCUUGCUAAUUUUUUG------------G--------UUU------UU (((((((((.....(((((.....))))).........(((((.(((((.....)))))....))))).....)))))))))...------------.--------...------.. ( -18.40) >DroWil_CAF1 4626 84 + 1 ACAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAU-------AUCGACAACAGACUUAGUUUUUUUUUUUUG------------G--------UUUU-----U- .(((..((..((((..(((...(((((....))))).(((.((((...-------...)))))))))).)))).....))..)))------------.--------....-----.- ( -13.90) >DroPer_CAF1 2785 96 + 1 AUAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACUUAUCGACAACAGACUU-UUGCUAAUUUUUUG------------G--------GUUUCCUUUCU .(((..(((((.((..(((...(((((....))))).(((.((((.(((.....))).))))))))))..-.)).)))))..)))------------.--------........... ( -14.70) >consensus AAAAUUAAUUAUUAUCGUCAAUGUGACGAAAGUUAUUUGUCUGUCAAUAAUACUUAUCGACAACAGACUU_UUGCUAAUUUUUUG____________G________UUUU____UUU ..............(((((.....)))))(((((..(((((.................)))))..)))))............................................... (-11.70 = -11.70 + -0.00)
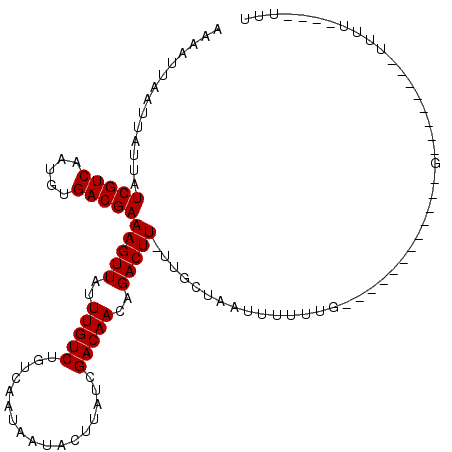
| Location | 9,793,445 – 9,793,538 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -17.63 |
| Consensus MFE | -15.49 |
| Energy contribution | -15.32 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975460 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
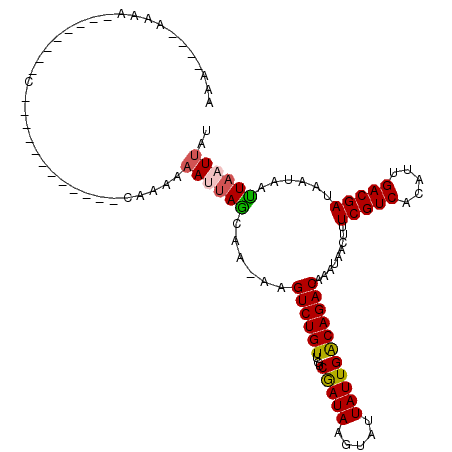
>2L_DroMel_CAF1 9793445 93 - 22407834 CAA----AAAG--------C------------AAAAAAAUUAGCAAGAAGUCUGUUGUCAAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUU ...----....--------.------------...((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......)))))))) ( -19.70) >DroVir_CAF1 2730 109 - 1 CAA----AAAACAAAAAAAAACCCAAAUUAGCAA---CA-UAAAAAAAAGUCUGUUGUCGAUACGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUGU ...----..................((((((...---..-.........((((((((..(((....)))..)))))))).........(((((.....)))))......)))))).. ( -16.40) >DroPse_CAF1 2785 96 - 1 AGAAAGGAAAC--------C------------CAAAAAAUUAGCAA-AAGUCUGUUGUCGAUAAGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUAU .....((....--------)------------)....((((((...-..((((((((..(((....)))..)))))))).........(((((.....)))))......)))))).. ( -17.50) >DroEre_CAF1 5040 91 - 1 AA------AAA--------C------------CAAAAAAUUAGCAAGAAGUCUGUUGUCAAUAAGAAUUAUUGGCAGACAAAUAACUUUCGUCACAUUGACGAUAAUAACUAAUUUU ..------...--------.------------...((((((((......((((((...(((((.....))))))))))).........(((((.....)))))......)))))))) ( -19.70) >DroWil_CAF1 4626 84 - 1 -A-----AAAA--------C------------CAAAAAAAAAAAACUAAGUCUGUUGUCGAU-------AUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUGU -.-----....--------.------------.................((((((((.....-------..)))))))).........(((((.....))))).............. ( -15.00) >DroPer_CAF1 2785 96 - 1 AGAAAGGAAAC--------C------------CAAAAAAUUAGCAA-AAGUCUGUUGUCGAUAAGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUAU .....((....--------)------------)....((((((...-..((((((((..(((....)))..)))))))).........(((((.....)))))......)))))).. ( -17.50) >consensus AAA____AAAA________C____________CAAAAAAUUAGCAA_AAGUCUGUUGUCGAUAAGUAUUAUUGACAGACAAAUAACUUUCGUCACAUUGACGAUAAUAAUUAAUUAU .....................................((((((......((((((...(((((.....))))))))))).........(((((.....)))))......)))))).. (-15.49 = -15.32 + -0.18)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:51:02 2006