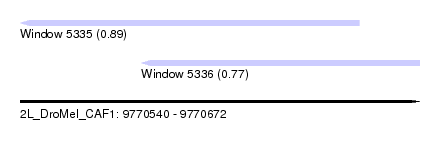
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,770,540 – 9,770,672 |
| Length | 132 |
| Max. P | 0.885334 |

| Location | 9,770,540 – 9,770,652 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 77.22 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -15.55 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.885334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
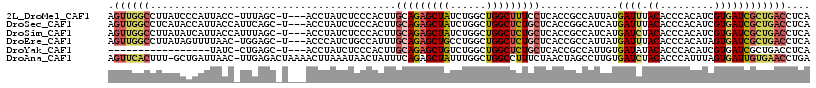
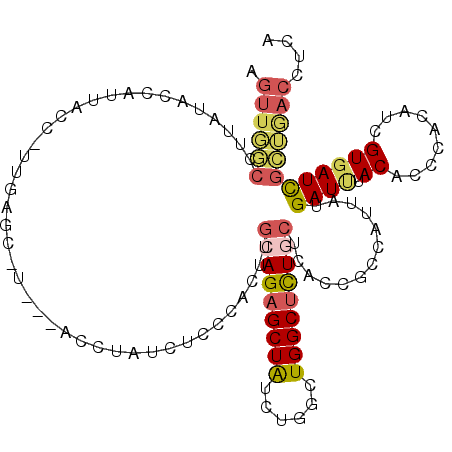
>2L_DroMel_CAF1 9770540 112 - 22407834 AGUUGGCCUUAUCCCAUUACC-UUUAGC-U---ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUUUCCUCACCGCCAUUAUGAUUUACACCCACAUCGUGAUCGCUGACCUCA (((..(((.............-....((-.---...............))(((....))))))..))).........((.((((((((..........)))))))).))........ ( -18.89) >DroSec_CAF1 52586 113 - 1 AGUUGGCCUCAUACCAUUACCAUUCAGC-U---ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUCUGCUCACCGGCAUCAUGAUUUACACCCACAUCGUGAUCGCUGACCUCA ((((((.................)))))-)---...............(((((((((......)))))))))....((((((((((((..........)))))))).))))...... ( -28.93) >DroSim_CAF1 52038 113 - 1 AGUUGGCCUUAUAUCAUUACCAUUUAGC-U---ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUCUGCUCACCGCCAUCAUGAUCUACACCCACAUCGUGAUCGCUGACCUCA .((..((.....................-.---...............(((((((((......)))))))))........((((((((..........)))))))).))..)).... ( -26.70) >DroEre_CAF1 52031 112 - 1 AGUUGGCCUUAUAGUUUUAAC-UGGAGC-U---ACCCAUCUGCCAUUUGCAGAGCUGCCUGGCUGGCUCUGCUCACCGCCAUUAUGAUUUACACCCACAUAGUGAUCGCUGACCUCA .((..((....(((((((...-.)))))-)---).......((.....((((((((((...)).)))))))).....))(((((((...........)))))))...))..)).... ( -29.50) >DroYak_CAF1 54192 95 - 1 -----------------UAUC-CUGAGC-U---ACCUAUCUCCCACUUGCAGAGCUGUCUGGCUGGCUCUGCUCACCGCCAUUGUGAUAUACACCCACAUCGUGAUCGCUGACCUCA -----------------....-..(((.-.---...............((((((((........)))))))).....((.((..((((..........))))..)).))....))). ( -22.30) >DroAna_CAF1 59949 115 - 1 AGUUCACUUU-GCUGAUUAAC-UUGAGACUAAAACUUAAAUAACUAUUUCAGAGCUAUUUGGCUGGCCUUUCUAACUAGCCUUGUGAUCUACACCCAUUUAGUGAUUGUGAACCUGA .((((((.(.-.((((.....-(((((.......)))))...........(((..(((..((((((.........))))))..))).)))........))))..)..)))))).... ( -25.30) >consensus AGUUGGCCUUAUACCAUUACC_UUGAGC_U___ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUCUGCUCACCGCCAUUAUGAUUUACACCCACAUCGUGAUCGCUGACCUCA .((((((.........................................(((((((((......))))))))).............((((.((.........)))))))))))).... (-15.55 = -16.08 + 0.53)
| Location | 9,770,580 – 9,770,672 |
|---|---|
| Length | 92 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 63.38 |
| Mean single sequence MFE | -18.44 |
| Consensus MFE | -9.76 |
| Energy contribution | -10.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.769424 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9770580 92 - 22407834 UCCAACUCAA-------UG------------UUUAAAAAAGUUGGCCUUAUCCCAUUACC-UUUAGC-U---ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUUUCCUCACC ..........-------..------------......(((((..(((.............-....((-.---...............))(((....))))))..)))))....... ( -13.89) >DroSec_CAF1 52626 100 - 1 CCUAACAUAAAGAUACCUG------------CUUAAAAAAGUUGGCCUCAUACCAUUACCAUUCAGC-U---ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUCUGCUCACC ..........(((((...(------------((.........(((.......))).........)))-.---...))))).......(((((((((......)))))))))..... ( -21.07) >DroSim_CAF1 52078 100 - 1 CCUAACAUAAAGAUACCUG------------UUUAAAACAGUUGGCCUUAUAUCAUUACCAUUUAGC-U---ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUCUGCUCACC ..........(((((.(((------------(.....)))).((((...................))-)---)..))))).......(((((((((......)))))))))..... ( -19.51) >DroEre_CAF1 52071 111 - 1 CUCAUUGGUCACAUUCCUACUGCUCCUAUUCUGUAAACCAGUUGGCCUUAUAGUUUUAAC-UGGAGC-U---ACCCAUCUGCCAUUUGCAGAGCUGCCUGGCUGGCUCUGCUCACC .....((((............(((((.(..((((((.((....))..))))))..)....-.)))))-.---........))))...((((((((((...)).))))))))..... ( -29.10) >DroAna_CAF1 59989 80 - 1 ----------------------------------AUAGUAGUUCACUUU-GCUGAUUAAC-UUGAGACUAAAACUUAAAUAACUAUUUCAGAGCUAUUUGGCUGGCCUUUCUAACU ----------------------------------.(((.((..(.....-.((((.....-(((((.......))))).........))))((((....)))))..))..)))... ( -8.64) >consensus CCCAACAUAAA_AU_CCUG____________UUUAAAAAAGUUGGCCUUAUACCAUUACC_UUGAGC_U___ACCUAUCUCCCACUUGCAGAGCUAUCUGGCUGGCUCUGCUCACC .......................................................................................(((((((((......)))))))))..... ( -9.76 = -10.16 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:49 2006