
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
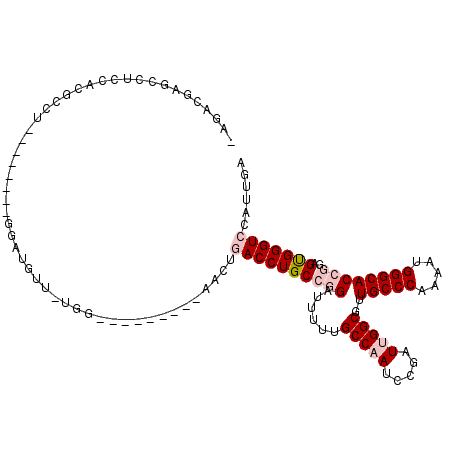
| Location | 9,749,824 – 9,749,923 |
| Length | 99 |
| Max. P | 0.696394 |

| Location | 9,749,824 – 9,749,923 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -22.79 |
| Energy contribution | -23.90 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627683 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
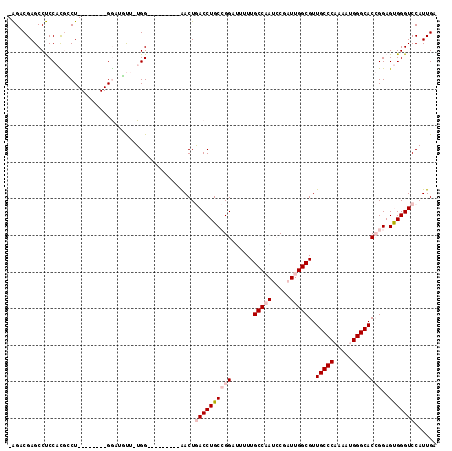
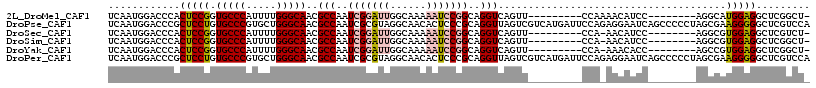
>2L_DroMel_CAF1 9749824 99 + 22407834 -AGCCGAGCCUCCAUGCCU--------GGAUGUUUUGG---------AACUGACCUGCCGGAUUUUUGCCAAUCCGAUUGGCGUUGCCCAAAAUGGGCACCGGAGUGGGUCCAUUGA -..((((((.((((....)--------))).).)))))---------....(((((.((((......((((((...))))))..(((((.....)))))))))...)))))...... ( -37.30) >DroPse_CAF1 32448 117 + 1 UGGACGAGCCCCCUUCGCUAGGGGGCUGAUUCCUCUGGAAUCAUGACGACUAACCUGCGGGAGUGUUGCCUACGCGAUUGGCGUUGCCCAGCACGGGCACAGGAGCGGGUCCAUUGA (((((.((((((((.....))))))))((((((...))))))....((.((..((((.(...((((((...((((.....))))....))))))...).)))))))).))))).... ( -49.90) >DroSec_CAF1 31932 98 + 1 -AGACGAGCCUCCACGCCU--------GGAUGUU-UGG---------AACUGACCUGCCGGAUUUUUGCCAAUCCGAUUGGCGUUGCCCAAAAUGGGCACCGGAGUGGGUCCAUUGA -...(((((.((((....)--------))).)))-)).---------....(((((.((((......((((((...))))))..(((((.....)))))))))...)))))...... ( -38.20) >DroSim_CAF1 30730 98 + 1 -AGCCGAGCCUCCACGCCU--------GGAUGUU-UGG---------AACUGACCUGCCGGAUUUUUGCCAAUCCGAUUGGCGUUGCCCAAAAUGGGCACCGGAGUGGGUCCAUUGA -..((((((.((((....)--------))).)))-)))---------....(((((.((((......((((((...))))))..(((((.....)))))))))...)))))...... ( -41.00) >DroYak_CAF1 33152 98 + 1 -AGCCGAGCCUCCACGGCU--------GGUGUUU-UGG---------AACUGACCUGCCGGAUUUUUGCCAAUCCGAUUGGCGUUGCCCAAAAUGGGCACCGGAGUGGGUCCAUUGA -.(((.((((.....))))--------)))....-(((---------(.((.((...((((......((((((...))))))..(((((.....))))))))).)).)))))).... ( -39.60) >DroPer_CAF1 31879 117 + 1 UGGACGAGCCCCCUUCGCUAGGGGGCUGAUUCCUCUGGAAUCAUGACGACUAACCUGCGGGAGUGUUGCCUACGCGAUUGGCGUUGCCCAGCACGGGCACAGGAGCGGGUCCAUUGA (((((.((((((((.....))))))))((((((...))))))....((.((..((((.(...((((((...((((.....))))....))))))...).)))))))).))))).... ( -49.90) >consensus _AGACGAGCCUCCACGCCU________GGAUGUU_UGG_________AACUGACCUGCCGGAUUUUUGCCAAUCCGAUUGGCGUUGCCCAAAAUGGGCACCGGAGUGGGUCCAUUGA ...................................................((((((((((......(((((.....)))))..(((((.....))))))))..)))))))...... (-22.79 = -23.90 + 1.11)

| Location | 9,749,824 – 9,749,923 |
|---|---|
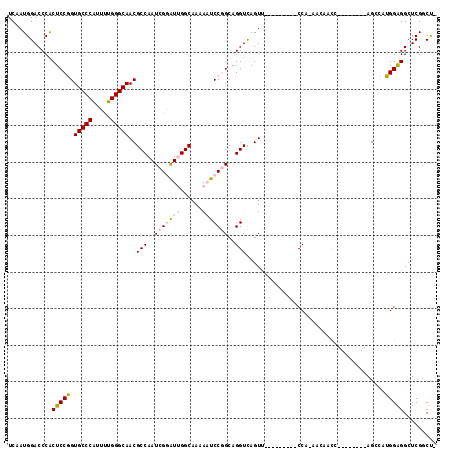
| Length | 99 |
| Sequences | 6 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Mean single sequence MFE | -38.92 |
| Consensus MFE | -22.04 |
| Energy contribution | -22.48 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696394 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9749824 99 - 22407834 UCAAUGGACCCACUCCGGUGCCCAUUUUGGGCAACGCCAAUCGGAUUGGCAAAAAUCCGGCAGGUCAGUU---------CCAAAACAUCC--------AGGCAUGGAGGCUCGGCU- .....((.((..(((((.(((((.....))))).)(((..(((((((......)))))))..((......---------)).........--------.)))..))))....))))- ( -32.10) >DroPse_CAF1 32448 117 - 1 UCAAUGGACCCGCUCCUGUGCCCGUGCUGGGCAACGCCAAUCGCGUAGGCAACACUCCCGCAGGUUAGUCGUCAUGAUUCCAGAGGAAUCAGCCCCCUAGCGAAGGGGGCUCGUCCA ....(((((.(((((((((((((.....)))).((((.....)))).(....)......)))))..)).))...(((((((...)))))))(((((((.....)))))))..))))) ( -47.40) >DroSec_CAF1 31932 98 - 1 UCAAUGGACCCACUCCGGUGCCCAUUUUGGGCAACGCCAAUCGGAUUGGCAAAAAUCCGGCAGGUCAGUU---------CCA-AACAUCC--------AGGCGUGGAGGCUCGUCU- .....((((...((((..(((((.....)))))(((((..(((((((......)))))))..((......---------)).-.......--------.)))))))))....))))- ( -36.80) >DroSim_CAF1 30730 98 - 1 UCAAUGGACCCACUCCGGUGCCCAUUUUGGGCAACGCCAAUCGGAUUGGCAAAAAUCCGGCAGGUCAGUU---------CCA-AACAUCC--------AGGCGUGGAGGCUCGGCU- .....((.((..((((..(((((.....)))))(((((..(((((((......)))))))..((......---------)).-.......--------.)))))))))....))))- ( -34.80) >DroYak_CAF1 33152 98 - 1 UCAAUGGACCCACUCCGGUGCCCAUUUUGGGCAACGCCAAUCGGAUUGGCAAAAAUCCGGCAGGUCAGUU---------CCA-AAACACC--------AGCCGUGGAGGCUCGGCU- ......((((....(((((((((.....)))))..((((((...))))))......))))..))))....---------...-.....((--------((((.....)))).))..- ( -35.00) >DroPer_CAF1 31879 117 - 1 UCAAUGGACCCGCUCCUGUGCCCGUGCUGGGCAACGCCAAUCGCGUAGGCAACACUCCCGCAGGUUAGUCGUCAUGAUUCCAGAGGAAUCAGCCCCCUAGCGAAGGGGGCUCGUCCA ....(((((.(((((((((((((.....)))).((((.....)))).(....)......)))))..)).))...(((((((...)))))))(((((((.....)))))))..))))) ( -47.40) >consensus UCAAUGGACCCACUCCGGUGCCCAUUUUGGGCAACGCCAAUCGGAUUGGCAAAAAUCCGGCAGGUCAGUU_________CCA_AACAACC________AGCCAUGGAGGCUCGGCU_ ............(((((.(((((.....)))))..(((..(((((((......)))))))..)))......................................)))))......... (-22.04 = -22.48 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:39 2006