
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,720,958 – 9,721,062 |
| Length | 104 |
| Max. P | 0.995486 |

| Location | 9,720,958 – 9,721,062 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.82 |
| Mean single sequence MFE | -31.97 |
| Consensus MFE | -12.95 |
| Energy contribution | -18.20 |
| Covariance contribution | 5.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.69 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.995486 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
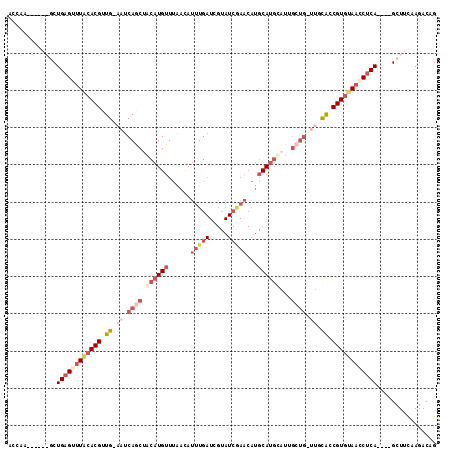
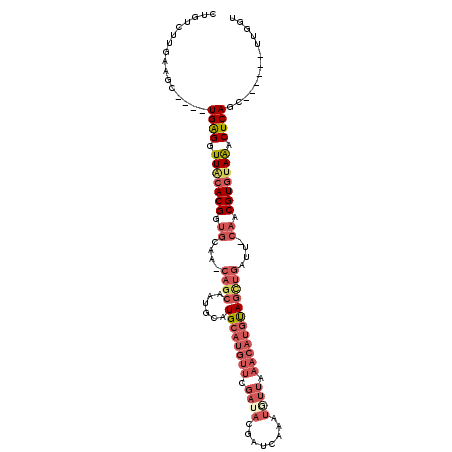
>2L_DroMel_CAF1 9720958 104 + 22407834 ACCAA-------CUGAGUUUGCACGUUGGAAUCAGCUACAUGUUUAAUAUUUGAUCGUAUCGAACAUGCAUGCAUUGCUGCUUGCAUCGUGUAACCUCA----GCUUCAAGACAG .....-------(((((.(((((((.((.((.((((..((((((..((((......))))..))))))........)))).)).)).))))))).))))----)........... ( -31.70) >DroSec_CAF1 3310 109 + 1 ACCAACAUAGAGCUGAGUUUACACGGUG-AAUCAACUACAUGUUUAACAUUUGAUCGUAUCAAACAUGCAUGUAUUGCUG-UUGCACCGUGUAAACUCA----GCUUCAAGACAG .........(((((((((((((((((((-((((((.(((((((......((((((...))))))...))))))))))..)-)).)))))))))))))))----))))........ ( -43.00) >DroSim_CAF1 3307 98 + 1 ACCAACAUAGAGCUGAGUUUACACGUUG-AAUCAGCUACAUGUUUAAAAUUUGAUCGUAUCGAACAUGCAU------------GCACCGUGUAACCUCA----GCUUCAAGACAG .........((((((((.((((((((((-...))))..(((((......((((((...))))))...))))------------)....)))))).))))----))))........ ( -26.40) >DroEre_CAF1 3370 87 + 1 -----------ACUGAGUCUACACGAUG-AAUCAGCUGCAUGUUAGGCAUUUG---------------CACGCCUUGCUG-ACUUGGCGUCCAAGCCCAGUCAGUUUUCAGACAG -----------.((((.((........)-).)))).....((((((((.....---------------...)))).((((-(((.(((......))).))))))).....)))). ( -26.80) >consensus ACCAA______GCUGAGUUUACACGUUG_AAUCAGCUACAUGUUUAACAUUUGAUCGUAUCGAACAUGCAUGCAUUGCUG_UUGCACCGUGUAACCUCA____GCUUCAAGACAG .............((((((((((((.((.((.((((.((((((......(((((.....)))))...))))))...)))).)).)).))))))))))))................ (-12.95 = -18.20 + 5.25)


| Location | 9,720,958 – 9,721,062 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.82 |
| Mean single sequence MFE | -33.63 |
| Consensus MFE | -13.20 |
| Energy contribution | -17.07 |
| Covariance contribution | 3.88 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.49 |
| Structure conservation index | 0.39 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990868 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
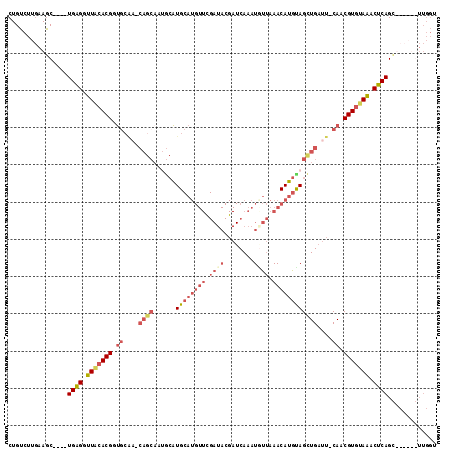
>2L_DroMel_CAF1 9720958 104 - 22407834 CUGUCUUGAAGC----UGAGGUUACACGAUGCAAGCAGCAAUGCAUGCAUGUUCGAUACGAUCAAAUAUUAAACAUGUAGCUGAUUCCAACGUGCAAACUCAG-------UUGGU .........(((----((((....((((.((....((((......((((((((..(((........)))..))))))))))))....)).))))....)))))-------))... ( -29.60) >DroSec_CAF1 3310 109 - 1 CUGUCUUGAAGC----UGAGUUUACACGGUGCAA-CAGCAAUACAUGCAUGUUUGAUACGAUCAAAUGUUAAACAUGUAGUUGAUU-CACCGUGUAAACUCAGCUCUAUGUUGGU .........(((----(((((((((((((((...-((((......(((((((((((((........)))))))))))))))))...-)))))))))))))))))).......... ( -46.20) >DroSim_CAF1 3307 98 - 1 CUGUCUUGAAGC----UGAGGUUACACGGUGC------------AUGCAUGUUCGAUACGAUCAAAUUUUAAACAUGUAGCUGAUU-CAACGUGUAAACUCAGCUCUAUGUUGGU .........(((----((((.(((((((..((------------.((((((((..(............)..)))))))))).....-...))))))).))))))).......... ( -28.90) >DroEre_CAF1 3370 87 - 1 CUGUCUGAAAACUGACUGGGCUUGGACGCCAAGU-CAGCAAGGCGUG---------------CAAAUGCCUAACAUGCAGCUGAUU-CAUCGUGUAGACUCAGU----------- ((((.((....((((((.(((......))).)))-)))..((((((.---------------...))))))..)).))))((((.(-(........)).)))).----------- ( -29.80) >consensus CUGUCUUGAAGC____UGAGGUUACACGGUGCAA_CAGCAAUGCAUGCAUGUUCGAUACGAUCAAAUGUUAAACAUGUAGCUGAUU_CAACGUGUAAACUCAGC______UUGGU ................((((.(((((((.((....((((......((((((((.((((........)))).))))))))))))....)).))))))).))))............. (-13.20 = -17.07 + 3.88)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:30 2006