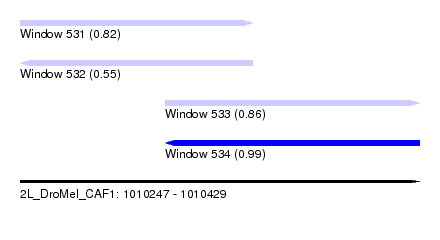
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,010,247 – 1,010,429 |
| Length | 182 |
| Max. P | 0.990986 |

| Location | 1,010,247 – 1,010,353 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 99.37 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -19.69 |
| Energy contribution | -19.47 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1010247 106 + 22407834 GCUUGAUUUAUUUUCAGUAAUCAUUUAAGUUCCUUUUUGAUUUAUCAGUUUAGUACAUUUAGAUAUUAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACC .............(((.((((.((((((((..((..((((....))))...))...)))))))))))).)))....((((((((........))))))))...... ( -20.30) >DroSec_CAF1 121975 106 + 1 GCUUGAUUUAUUUUCAGUAAUCAUUUAAGUUCCCUUUUGAUUUAUCAGUUUAGUACAUUUAGAUAUUAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACC .............(((.((((.((((((((...((.((((....))))...))...)))))))))))).)))....((((((((........))))))))...... ( -20.20) >DroSim_CAF1 122539 106 + 1 GCUUGAUUUAUUUUCAGUAAUCAUUUAAGUUCCCUUUUGAUUUAUCAGUUUAGUACAUUUAGAUAUUAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACC .............(((.((((.((((((((...((.((((....))))...))...)))))))))))).)))....((((((((........))))))))...... ( -20.20) >consensus GCUUGAUUUAUUUUCAGUAAUCAUUUAAGUUCCCUUUUGAUUUAUCAGUUUAGUACAUUUAGAUAUUAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACC .............(((.((((.((((((((...((.((((....))))...))...)))))))))))).)))....((((((((........))))))))...... (-19.69 = -19.47 + -0.22)

| Location | 1,010,247 – 1,010,353 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 99.37 |
| Mean single sequence MFE | -16.79 |
| Consensus MFE | -15.43 |
| Energy contribution | -15.76 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.92 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545887 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1010247 106 - 22407834 GGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUAAUAUCUAAAUGUACUAAACUGAUAAAUCAAAAAGGAACUUAAAUGAUUACUGAAAAUAAAUCAAGC (((.....((((((........)))))).....))).....((((...............))))...................(((((..(....)..)))))... ( -14.86) >DroSec_CAF1 121975 106 - 1 GGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUAAUAUCUAAAUGUACUAAACUGAUAAAUCAAAAGGGAACUUAAAUGAUUACUGAAAAUAAAUCAAGC (((.....((((((........)))))).....))).....((((...............))))(((((.(((....)))...))))).................. ( -17.76) >DroSim_CAF1 122539 106 - 1 GGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUAAUAUCUAAAUGUACUAAACUGAUAAAUCAAAAGGGAACUUAAAUGAUUACUGAAAAUAAAUCAAGC (((.....((((((........)))))).....))).....((((...............))))(((((.(((....)))...))))).................. ( -17.76) >consensus GGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUAAUAUCUAAAUGUACUAAACUGAUAAAUCAAAAGGGAACUUAAAUGAUUACUGAAAAUAAAUCAAGC (((.....((((((........)))))).....))).....((((...............))))(((((.(((....)))...))))).................. (-15.43 = -15.76 + 0.33)

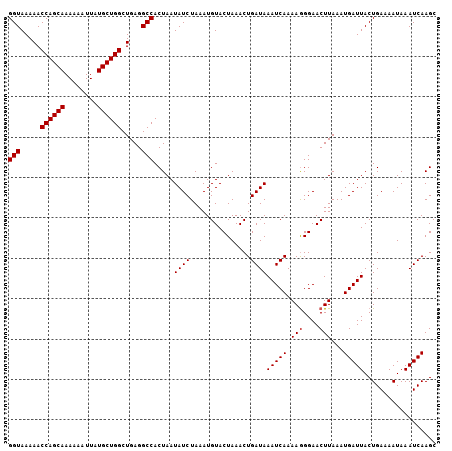
| Location | 1,010,313 – 1,010,429 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -18.30 |
| Consensus MFE | -16.42 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.90 |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.862207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1010313 116 + 22407834 UAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACCUUUUUUUUUGGAUUUUGGAUUUCUCAUCGGUUUUUUCGUCUUU----UUCAUCUUUGCUUAAUAACACAAAUCUUUGCCA ...((((...((((((((........))))))))...............((((((((((....))..(((.....))).....----..................)).))))))..)))) ( -18.70) >DroSec_CAF1 122041 115 + 1 UAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACCUUUU-UUUUGGAUUUUGGAUUUCUCAUUGGUUUUUUCGUCUUU---UUUCAUCU-UGCUUAAUAACACAAAUCUUUGCCA ...((((...((((((((........))))))))..........-....((((...(((..((.....))..)))..))))..---........-.....................)))) ( -18.00) >DroSim_CAF1 122605 113 + 1 UAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACCUUUU-UUUUGGAUUUUGGAUUU--CAUUGGUUUUUUCGUCUUU---UUUCAUCU-UGCUUAAUAACACAAAUCUUUGCCA ...((((...((((((((........))))))))....((....-....)).....((((((--.....(((.((..((....---........-.))..)).)))..))))))..)))) ( -18.02) >DroYak_CAF1 129254 112 + 1 UAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACCUUUU-UUUUGG-------GUUUCUUAUUAGUUUUUUCAUUUUUGCUUUUCAUCUUUGCUUAAUAACACAAAUCUUUGCCA ...((((...((((((((........))))))))...((((...-....))-------))........................................................)))) ( -18.50) >consensus UAGUGGCCUCAGCCAGCAUAAUUUUUUGCUGGUUUUUACCUUUU_UUUUGGAUUUUGGAUUUCUCAUUGGUUUUUUCGUCUUU___UUUCAUCU_UGCUUAAUAACACAAAUCUUUGCCA ...((((...((((((((........))))))))....((.........)).................................................................)))) (-16.42 = -16.43 + 0.00)

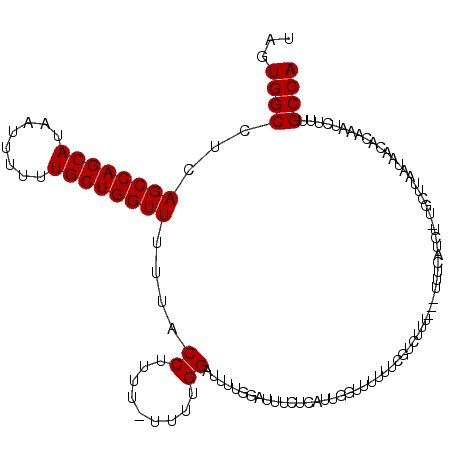
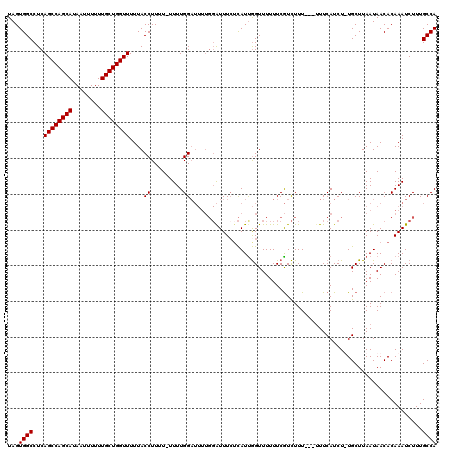
| Location | 1,010,313 – 1,010,429 |
|---|---|
| Length | 116 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.95 |
| Mean single sequence MFE | -18.32 |
| Consensus MFE | -17.40 |
| Energy contribution | -17.40 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.95 |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.990986 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1010313 116 - 22407834 UGGCAAAGAUUUGUGUUAUUAAGCAAAGAUGAA----AAAGACGAAAAAACCGAUGAGAAAUCCAAAAUCCAAAAAAAAAGGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUA ((((...(((((.((((....))))........----..................((....))..)))))..................((((((........)))))).....))))... ( -18.00) >DroSec_CAF1 122041 115 - 1 UGGCAAAGAUUUGUGUUAUUAAGCA-AGAUGAAA---AAAGACGAAAAAACCAAUGAGAAAUCCAAAAUCCAAAA-AAAAGGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUA ((((...(((((.((((....))))-........---..................((....))..))))).....-............((((((........)))))).....))))... ( -18.00) >DroSim_CAF1 122605 113 - 1 UGGCAAAGAUUUGUGUUAUUAAGCA-AGAUGAAA---AAAGACGAAAAAACCAAUG--AAAUCCAAAAUCCAAAA-AAAAGGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUA ((((..((.(((((........)))-))......---...................--.................-............((((((........))))))))...))))... ( -17.30) >DroYak_CAF1 129254 112 - 1 UGGCAAAGAUUUGUGUUAUUAAGCAAAGAUGAAAAGCAAAAAUGAAAAAACUAAUAAGAAAC-------CCAAAA-AAAAGGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUA ((((..((.(((((.(((((.......)))))...)))))....................((-------(.....-....))).....((((((........))))))))...))))... ( -20.00) >consensus UGGCAAAGAUUUGUGUUAUUAAGCA_AGAUGAAA___AAAGACGAAAAAACCAAUGAGAAAUCCAAAAUCCAAAA_AAAAGGUAAAAACCAGCAAAAAAUUAUGCUGGCUGAGGCCACUA ((((..((((((.((((....)))).))))..........................................................((((((........))))))))...))))... (-17.40 = -17.40 + 0.00)

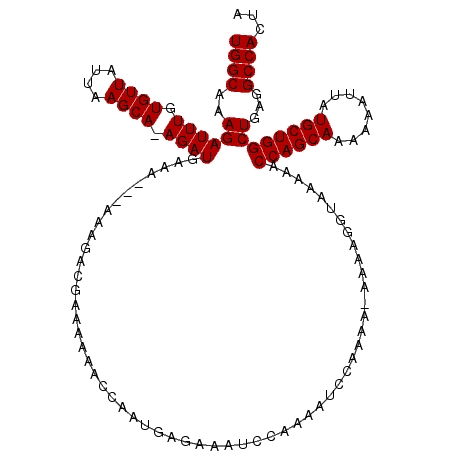
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:19 2006