
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,711,185 – 9,711,314 |
| Length | 129 |
| Max. P | 0.957633 |

| Location | 9,711,185 – 9,711,277 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 84.23 |
| Mean single sequence MFE | -21.70 |
| Consensus MFE | -16.25 |
| Energy contribution | -16.42 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.728958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9711185 92 - 22407834 GGAACUGGUCACAC--UUCCUUUCCACGCGAAACCAACAACACAAAAAACGUGUGUUUUGGUC-AACAGCGUGUUUUCUUUAAAAUAAAUCUGUA----- ((((.((....)).--))))....(((((...(((((.((((((.......))))))))))).-....)))))......................----- ( -22.60) >DroSec_CAF1 2156 92 - 1 UGAACUGGUCACAC--UUCCUUUCCACGCGAGACCAACAACACAAAAAACGUGUGUUUUGGUC-AACAGCGUGUUUUCUUUAAAAUAAAUCUGUA----- .(((.((....)).--))).....(((((..((((((.((((((.......))))))))))))-....)))))......................----- ( -22.30) >DroSim_CAF1 2149 92 - 1 UGAACUGGUCACAC--UUCCUUUCCACGCGAGACCAACAACACAAAAAACGUGUGUUUUGGUC-AACAGCGUGUUUUCUUUAAAAUAAAUCUGUA----- .(((.((....)).--))).....(((((..((((((.((((((.......))))))))))))-....)))))......................----- ( -22.30) >DroEre_CAF1 2156 92 - 1 CAAGCCGGUCACAC--UUCCUUUCCACGCGAAACCAACAACACAAAAAACGUGUGUUUUGGUG-AACAGCGUGUUUUCCUUUAAAUAAAUCUGUU----- ......((......--..))....(((((...(((((.((((((.......))))))))))).-....)))))......................----- ( -19.40) >DroYak_CAF1 2161 92 - 1 CAAAGCGGCCACAC--UUCCUUUCCACGCUGAACCAACAACACAAAAAACGUGUGUUUUGGUC-AACAGCGUGUUUUCCUUUAAAUAAAUCUGUA----- .((((.((......--..))....(((((((.(((((.((((((.......))))))))))).-..))))))).....)))).............----- ( -21.50) >DroMoj_CAF1 2265 97 - 1 GUGAACGGUCACAUCGUUCCUUUCCACGC--GACCAACAACACAA-AAACGUGUGUUUUGGCUACGGUGCGUGUUCUUAAUAAAAUAAAACAUUUCGGAA ..(((((((...)))))))..((((((((--(.((((.((((((.-.....))))))))))(....))))))(((.(((......)))))).....)))) ( -22.10) >consensus CGAACCGGUCACAC__UUCCUUUCCACGCGAAACCAACAACACAAAAAACGUGUGUUUUGGUC_AACAGCGUGUUUUCUUUAAAAUAAAUCUGUA_____ ........................(((((...(((((.((((((.......)))))))))))......)))))........................... (-16.25 = -16.42 + 0.17)


| Location | 9,711,212 – 9,711,314 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 77.88 |
| Mean single sequence MFE | -20.32 |
| Consensus MFE | -11.33 |
| Energy contribution | -11.97 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.56 |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.957633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
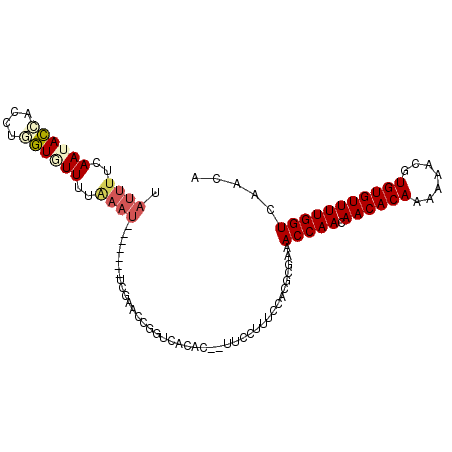
>2L_DroMel_CAF1 9711212 102 - 22407834 UAUUUUCAAUACCACCUGGUCUUUUAAAUAACAAUUUGGAACUGGUCACAC--UUCCUUUCCACGCGAAACCAACAACACAAAAAACGUGUGUUUUGGUCAACA (((((..((.(((....))).))..)))))......(((((..((......--..)).)))))......(((((.((((((.......)))))))))))..... ( -18.90) >DroVir_CAF1 2325 89 - 1 CAAUUUCAAUAUUUUGUAAUAUUG--U----------GUCUACGGUCACACUGUCCCUUCCCACGC--GACCAAUAACACAA-AAACGUGUGUUUUGGUUCGGU ...............(..(((.((--(----------(.(....).)))).)))..)......((.--((((((.((((((.-.....)))))))))))))).. ( -20.50) >DroSec_CAF1 2183 102 - 1 UAUUUUUAAUACCACCUCGUGUUUUAAAUGAAAACAUUGAACUGGUCACAC--UUCCUUUCCACGCGAGACCAACAACACAAAAAACGUGUGUUUUGGUCAACA ..........((((..(((((((((.....)))))).)))..)))).....--...............((((((.((((((.......)))))))))))).... ( -22.30) >DroSim_CAF1 2176 102 - 1 UAUUUUUAAUACCACCUCGUGUUUUAAAUAAAAACAUUGAACUGGUCACAC--UUCCUUUCCACGCGAGACCAACAACACAAAAAACGUGUGUUUUGGUCAACA ..........((((..(((((((((.....)))))).)))..)))).....--...............((((((.((((((.......)))))))))))).... ( -22.20) >DroEre_CAF1 2183 95 - 1 UAUUUUCAAUACCACCUGGUGUUUUGAAU-------UCAAGCCGGUCACAC--UUCCUUUCCACGCGAAACCAACAACACAAAAAACGUGUGUUUUGGUGAACA ....(((((((((....)))))..))))(-------((..((.((......--.......))..))...(((((.((((((.......)))))))))))))).. ( -17.82) >DroYak_CAF1 2188 95 - 1 UAUUUACAAAACCACCUGGUGUUUUGAAU-------UCAAAGCGGCCACAC--UUCCUUUCCACGCUGAACCAACAACACAAAAAACGUGUGUUUUGGUCAACA ................((((((((((...-------.)))))).))))...--...........(.(((.((((.((((((.......))))))))))))).). ( -20.20) >consensus UAUUUUCAAUACCACCUGGUGUUUUAAAU_______UCGAACCGGUCACAC__UUCCUUUCCACGCGAAACCAACAACACAAAAAACGUGUGUUUUGGUCAACA .((((..((((((....))))))..))))........................................(((((.((((((.......)))))))))))..... (-11.33 = -11.97 + 0.64)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:23 2006