
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
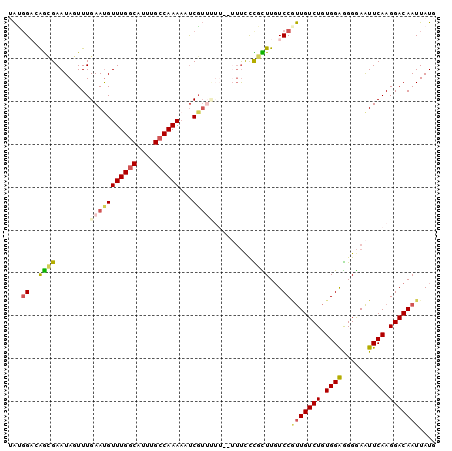
| Location | 9,666,579 – 9,666,677 |
| Length | 98 |
| Max. P | 0.804965 |

| Location | 9,666,579 – 9,666,677 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -28.72 |
| Consensus MFE | -16.50 |
| Energy contribution | -17.03 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.57 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.524046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9666579 98 + 22407834 UAUGGACAGCGAAUAGUUUGAAUGUUUGGCAUUUGCCAAAAAUCGUUUUU--UUUUCCGCUUUUCCGUUGUCUGUGGAGGGGAAUUCAAGGACAAUUAUG ...(((.((((((.((..(((...((((((....))))))..)))..)).--..)).))))..)))(((((((.((((......)))).))))))).... ( -26.30) >DroSec_CAF1 24861 97 + 1 UAUGGACAGCGAAUAGUUUGAAUGUUUGGCAUUUGCCAAAAAUCGUUUUU---UUUCCGCUUGUCCGUUGUCUGUGGAGGGGAAUUCAAGGACAAUUAUG ...((((((((((.((..(((...((((((....))))))..)))..)).---.)).))).)))))(((((((.((((......)))).))))))).... ( -30.50) >DroSim_CAF1 25864 99 + 1 AAUGGACAGCGAAUAGUUUGAAUGUUUGGCAUUUGCCAAAAAUCGUUUUUU-UUUUUCGCUUGUCCGUUGUCUGUGGAGGGGAAUUCAAGGACAAUUAUG ...((((((((((......(((((((((((....))))))...)))))...-...))))).)))))(((((((.((((......)))).))))))).... ( -34.40) >DroEre_CAF1 30499 95 + 1 UACUGCGAACGAAUAGUUUGAACGUUUGGCAUUUGCCAAAAUUCGU-----UUUUCCUGUUUGUCCCUUGUCUGUGGAAGGGAAUUCAAGGACAAUUAUG ......((((((((.((....)).((((((....))))))))))))-----)).((((.....((((((........)))))).....))))........ ( -26.50) >DroYak_CAF1 25010 100 + 1 UAUGGCGAGUGAAUAGUUUGAAUGUUUGGCAUUUGCCAAAAUUCGUUCUUUUUUUCCCGUUUGUCCAUUGUCUGUGGAGGGGAAUUCAAGGACAAUUAUG ............((((((.((((.((((((....))))))))))((((((..((((((.....(((((.....)))))))))))...)))))))))))). ( -28.30) >DroAna_CAF1 27680 93 + 1 AGGGGAGGGAGAAUAGUUUGAAUGUUUGCCAUUUGCCAAAUUCCC-------AACCCUGUUUGUCCGCUGUCUCUGGGGGGAAGUUCAAGGACAAUUAUA ((((..(((((....((..(((((.....)))))))....)))))-------..))))..(((((((((.((((....)))))))....))))))..... ( -26.30) >consensus UAUGGACAGCGAAUAGUUUGAAUGUUUGGCAUUUGCCAAAAAUCGUUUUU__UUUCCCGCUUGUCCGUUGUCUGUGGAGGGGAAUUCAAGGACAAUUAUG ...((..((((........(((((((((((....))))))...))))).........))))...))(((((((.((((......)))).))))))).... (-16.50 = -17.03 + 0.53)


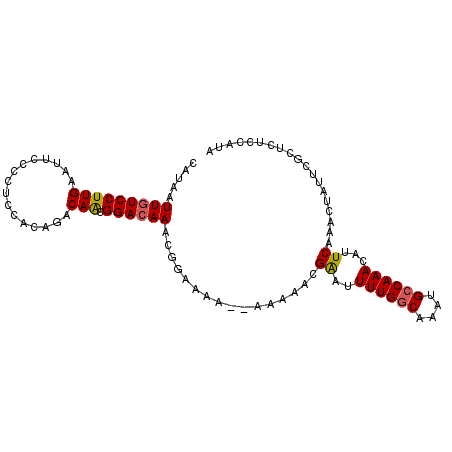
| Location | 9,666,579 – 9,666,677 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.02 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -12.60 |
| Energy contribution | -12.66 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.36 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.804965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9666579 98 - 22407834 CAUAAUUGUCCUUGAAUUCCCCUCCACAGACAACGGAAAAGCGGAAAA--AAAAACGAUUUUUGGCAAAUGCCAAACAUUCAAACUAUUCGCUGUCCAUA .....(((((..((..........))..))))).(((..((((((...--......(((.((((((....)))))).))).......)))))).)))... ( -20.69) >DroSec_CAF1 24861 97 - 1 CAUAAUUGUCCUUGAAUUCCCCUCCACAGACAACGGACAAGCGGAAA---AAAAACGAUUUUUGGCAAAUGCCAAACAUUCAAACUAUUCGCUGUCCAUA .....(((((..((..........))..))))).(((((.(((((..---......(((.((((((....)))))).))).......))))))))))... ( -24.96) >DroSim_CAF1 25864 99 - 1 CAUAAUUGUCCUUGAAUUCCCCUCCACAGACAACGGACAAGCGAAAAA-AAAAAACGAUUUUUGGCAAAUGCCAAACAUUCAAACUAUUCGCUGUCCAUU .....(((((..((..........))..))))).(((((.(((((...-.......(((.((((((....)))))).))).......))))))))))... ( -24.33) >DroEre_CAF1 30499 95 - 1 CAUAAUUGUCCUUGAAUUCCCUUCCACAGACAAGGGACAAACAGGAAAA-----ACGAAUUUUGGCAAAUGCCAAACGUUCAAACUAUUCGUUCGCAGUA ....(((((...(((((((((((........))))))............-----..((((((((((....)))))).)))).....)))))...))))). ( -22.40) >DroYak_CAF1 25010 100 - 1 CAUAAUUGUCCUUGAAUUCCCCUCCACAGACAAUGGACAAACGGGAAAAAAAGAACGAAUUUUGGCAAAUGCCAAACAUUCAAACUAUUCACUCGCCAUA .......((.(((...(((((.((((.......)))).....)))))...))).))((((((((((....)))))).))))................... ( -21.00) >DroAna_CAF1 27680 93 - 1 UAUAAUUGUCCUUGAACUUCCCCCCAGAGACAGCGGACAAACAGGGUU-------GGGAAUUUGGCAAAUGGCAAACAUUCAAACUAUUCUCCCUCCCCU .....(((((((((..(((.......))).))).))))))..((((..-------((((...(((..((((.....))))....)))...))))..)))) ( -22.80) >consensus CAUAAUUGUCCUUGAAUUCCCCUCCACAGACAACGGACAAACGGAAAA__AAAAACGAAUUUUGGCAAAUGCCAAACAUUCAAACUAUUCGCUCUCCAUA .....(((((((((................))).))))))................((..((((((....))))))...))................... (-12.60 = -12.66 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:05 2006