
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,664,783 – 9,664,916 |
| Length | 133 |
| Max. P | 0.563474 |

| Location | 9,664,783 – 9,664,893 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -12.49 |
| Energy contribution | -15.97 |
| Covariance contribution | 3.47 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.60 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.01 |
| SVM RNA-class probability | 0.536889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9664783 110 + 22407834 GCUUGCCAAAAGGAAAAUGGAGGCAGC--AU-AAAAGGAAACGGAAAAAGUUGCGAAUAAAUGGAAUUAUAUC-------CUUGGCAAUGGCUGCAGUCGAGCAGAAAUGAUCAACUGCU ((((((((..((((........(((((--..-....(....).......)))))..((((......)))).))-------)))))))..))).(((((.((.((....)).)).))))). ( -31.22) >DroPse_CAF1 44464 117 + 1 GCUUGCCAAAACUGAGAUAAAAAUCAC--GA-GAAAAGAGUGGGAAAACUUUGCAAAUAAAUGGAAUUAUUUCCAGGGGCCUUGACAGUGGCUGCACUCGAGCAGAAAUGAUCAACUGCA ((..(((...((((.(((....))).(--((-(...(((((......)))))((.......(((((....)))))...)))))).))))))).))......((((..........)))). ( -26.00) >DroSec_CAF1 23010 112 + 1 GCUUGCCAAAAGGAAAGUGGAGGCAGCGCCU-AAAAGGAAACGGAAAAAGUUGCGAAUAAAUGGAAUUAUAUC-------CUUGGCAAUGGCUGCAGUCGAGCAGAAAUGAUCAACUGCU ((((((((..((((........(((((....-....(....).......)))))..((((......)))).))-------)))))))..))).(((((.((.((....)).)).))))). ( -32.16) >DroYak_CAF1 23131 110 + 1 GCUUGCCAAAAGGAAAGUGGAGGCAGC--AU-GAAAGGAAACGGAAUAAGUUGCGAAUAAAUGGAAUUAUAUC-------CUUGGCAAUGGCUGCAGUCGAGCAGAAAUGAUCAACUGCU ((((((((..((((........(((((--..-....(....).......)))))..((((......)))).))-------)))))))..))).(((((.((.((....)).)).))))). ( -31.22) >DroAna_CAF1 25999 111 + 1 GCUUGCCAAACGGAAAGUGGAGGCCAA--ACAGAAAGGAAACGGAAAAAGUUGCGAAUAAAUGCAAUUAUAUC-------CUUGGCAAUGGCAGCAGUCGAGCAGAAAUGAUCAACUGUA ...(((((.((.....))....(((((--.......(....)(((.(.(((((((......))))))).).))-------))))))..)))))(((((.((.((....)).)).))))). ( -32.80) >DroPer_CAF1 44646 117 + 1 GCUUGCCAAAACUGAGAGAAAAAUCAA--GG-GAAAAGAGUGGGAAAACUUUGCAAAUAAAUGGAAUUAUUUCCAGGGGCCUUGACAGUGGCUGCACGCGAGCAGAAAUGAUCAACUGCA ((.(((....((((.........((((--((-....(((((......))))).........(((((....)))))....))))))))))....))).))..((((..........)))). ( -28.00) >consensus GCUUGCCAAAAGGAAAAUGGAGGCAAC__AU_GAAAGGAAACGGAAAAAGUUGCGAAUAAAUGGAAUUAUAUC_______CUUGGCAAUGGCUGCAGUCGAGCAGAAAUGAUCAACUGCA (((((((((...........................(....)......((((.((......)).)))).............))))))..))).(((((.((.((....)).)).))))). (-12.49 = -15.97 + 3.47)

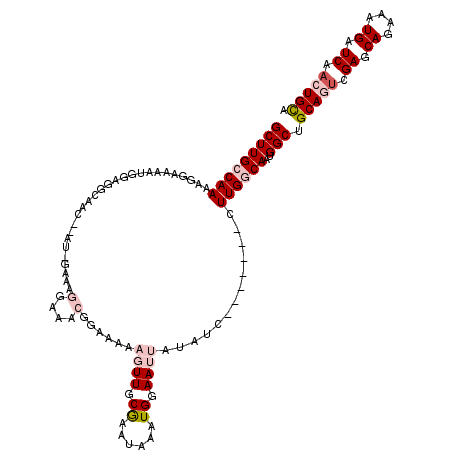
| Location | 9,664,783 – 9,664,893 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.27 |
| Mean single sequence MFE | -24.59 |
| Consensus MFE | -13.52 |
| Energy contribution | -13.80 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.563474 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9664783 110 - 22407834 AGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUUUUCCGUUUCCUUUU-AU--GCUGCCUCCAUUUUCCUUUUGGCAAGC .((((((((.((....)).)))))))).....((((((((-------((......)))........(((.(.................-..--).))).............))))))).. ( -25.31) >DroPse_CAF1 44464 117 - 1 UGCAGUUGAUCAUUUCUGCUCGAGUGCAGCCACUGUCAAGGCCCCUGGAAAUAAUUCCAUUUAUUUGCAAAGUUUUCCCACUCUUUUC-UC--GUGAUUUUUAUCUCAGUUUUGGCAAGC ((((.((((.((....)).)))).)))).....(((((((((...((.((((((......)))))).))...................-..--(.(((....))).).)))))))))... ( -23.90) >DroSec_CAF1 23010 112 - 1 AGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUUUUCCGUUUCCUUUU-AGGCGCUGCCUCCACUUUCCUUUUGGCAAGC .((((((((.((....)).)))))))).....((((((((-------((......)))..............................-((((...))))...........))))))).. ( -27.90) >DroYak_CAF1 23131 110 - 1 AGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUAUUCCGUUUCCUUUC-AU--GCUGCCUCCACUUUCCUUUUGGCAAGC .((((((((.((....)).)))))))).....((((((((-------((......)))........(((.(.................-..--).))).............))))))).. ( -25.31) >DroAna_CAF1 25999 111 - 1 UACAGUUGAUCAUUUCUGCUCGACUGCUGCCAUUGCCAAG-------GAUAUAAUUGCAUUUAUUCGCAACUUUUUCCGUUUCCUUUCUGU--UUGGCCUCCACUUUCCGUUUGGCAAGC ..(((((((.((....)).)))))))......((((((((-------((.....((((........)))).....))).............--.(((...)))........))))))).. ( -23.90) >DroPer_CAF1 44646 117 - 1 UGCAGUUGAUCAUUUCUGCUCGCGUGCAGCCACUGUCAAGGCCCCUGGAAAUAAUUCCAUUUAUUUGCAAAGUUUUCCCACUCUUUUC-CC--UUGAUUUUUCUCUCAGUUUUGGCAAGC .((((..........))))..((.(((....((((((((((....((.((((((......)))))).)).(((......)))......-))--)))).........))))....))).)) ( -21.20) >consensus AGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG_______GAUAUAAUUCCAUUUAUUCGCAACUUUUUCCGUUUCCUUUC_AU__GCGGCCUCCACUUUCCUUUUGGCAAGC .((((((((.((....)).)))))))).((..((((((((.......((.((((......)))))).............................((....)).......)))))))))) (-13.52 = -13.80 + 0.28)

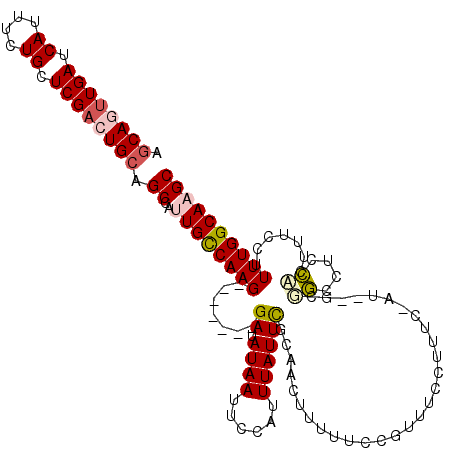
| Location | 9,664,820 – 9,664,916 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.10 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -14.52 |
| Energy contribution | -14.72 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9664820 96 - 22407834 CCUUCGUCGGU-----------------CGUCCUGCCUGCAGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUUUUCCGU .......(((.-----------------.....(((..((.((((((((.((....)).)))))))).)).........(-------((......)))........))).......))). ( -22.12) >DroPse_CAF1 44501 120 - 1 CCAUCAUCCGGCUGCUGGCUGCUGGCUGCCGCCUGCCGGCUGCAGUUGAUCAUUUCUGCUCGAGUGCAGCCACUGUCAAGGCCCCUGGAAAUAAUUCCAUUUAUUUGCAAAGUUUUCCCA .........((((..((((.((.(((....))).)).(((((((.((((.((....)).)))).)))))))...)))).))))..((.((((((......)))))).))........... ( -40.70) >DroSec_CAF1 23049 92 - 1 CCCUCGU---------------------CGUCCUGCCUGCAGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUUUUCCGU .......---------------------.(((((((..((.((((((((.((....)).)))))))).))....))..))-------))).............................. ( -19.70) >DroEre_CAF1 22843 92 - 1 CCCUCGU---------------------CGUCCUGCCUGCAGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUAUUCCGU .......---------------------.(((((((..((.((((((((.((....)).)))))))).))....))..))-------))).............................. ( -19.70) >DroYak_CAF1 23168 92 - 1 CCUUCGU---------------------CGUCCUGCCUGCAGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG-------GAUAUAAUUCCAUUUAUUCGCAACUUAUUCCGU .......---------------------.(((((((..((.((((((((.((....)).)))))))).))....))..))-------))).............................. ( -19.70) >DroAna_CAF1 26037 92 - 1 CCUUCCC---------------------AGUCCUGCCUGCUACAGUUGAUCAUUUCUGCUCGACUGCUGCCAUUGCCAAG-------GAUAUAAUUGCAUUUAUUCGCAACUUUUUCCGU .......---------------------.(((((((..((..(((((((.((....)).)))))))..))....))..))-------)))....((((........)))).......... ( -19.70) >consensus CCUUCGU_____________________CGUCCUGCCUGCAGCAGUUGAUCAUUUCUGCUCGACUGCAGCCAUUGCCAAG_______GAUAUAAUUCCAUUUAUUCGCAACUUUUUCCGU .................................(((..((.((((((((.((....)).)))))))).)).................((.((((......)))))))))........... (-14.52 = -14.72 + 0.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:50:03 2006