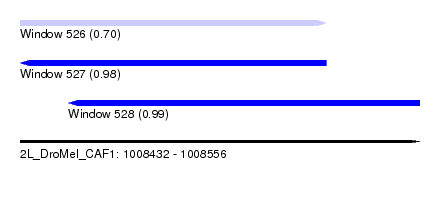
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 1,008,432 – 1,008,556 |
| Length | 124 |
| Max. P | 0.992194 |

| Location | 1,008,432 – 1,008,527 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -29.96 |
| Consensus MFE | -16.58 |
| Energy contribution | -17.86 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.697469 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1008432 95 + 22407834 UUGCACAU-------------UAACUGUGUGGUUAUUUCACUAGAUAAUUUGCUUUUGGCUUAGAAGCCAGGUCUCUUUGGUGAGACGAAACAAGUGGUUAUUGGCUU ..((.((.-------------(((((.(((..(..((((((((((......(((..(((((....))))))))...))))))))))..).)))...))))).)))).. ( -25.90) >DroSec_CAF1 120180 108 + 1 UUGCACAUUUCAAGUGUGGCUUAACUGUGUGGUUAUUUCGGUAGAUAUUCUGCUUUUGGCUUAGAAGCCAGGUCUCUUUGGUGAGACGAAACAAUUGGGUGGUGGCUU ...(((((.....)))))(((..((..(.(((((.(((((((((.....))))).((((((....))))))(((((......))))))))).))))).)..))))).. ( -33.10) >DroSim_CAF1 120754 108 + 1 UUGCACAUUUCAAGUGUGGCUUAAUUGUGUGGUUAUUUUGGUAGAUAUUUUGCAUUUGGCUUAGAAGCCAGGUCUCUUUGGUGAGACGAAACAAUUGGGUGGUGGCUU ...(((((.....)))))(((((((((((..(.(((((....)))))..)..)..((((((....))))))(((((......)))))...))))))))))........ ( -30.20) >DroEre_CAF1 125310 97 + 1 UUGCACAUUUCAA-UGUGGCUUAACUG--UGGUUAUUUCCAUAGAUAUUUGGCUAAGGGCUU-GGAGCCAGGUCUCUGCGCUGAGACGAAACAAGUGUGA-------U (..(((.((((..-..((((((..(((--(((......))))))......(((.....))).-.)))))).(((((......)))))))))...)))..)-------. ( -31.70) >DroYak_CAF1 127308 106 + 1 UUGCACAUUUCAAGUGUGGAUUAACUG--UGGUUAUUUCGCUAGAUAUUUUGCUAUUGGCUUAGGAGCCAGGUCUCUUUGCUGAGACGAAACAAUUGUGUAGUGACUC (((((((((((....(((((.((((..--..)))).)))))..............((((((....))))))(((((......)))))))))....)))))))...... ( -28.90) >consensus UUGCACAUUUCAAGUGUGGCUUAACUGUGUGGUUAUUUCGCUAGAUAUUUUGCUUUUGGCUUAGAAGCCAGGUCUCUUUGGUGAGACGAAACAAUUGGGUAGUGGCUU ...(((((.....)))))...(((((....)))))((((................((((((....))))))(((((......)))))))))................. (-16.58 = -17.86 + 1.28)

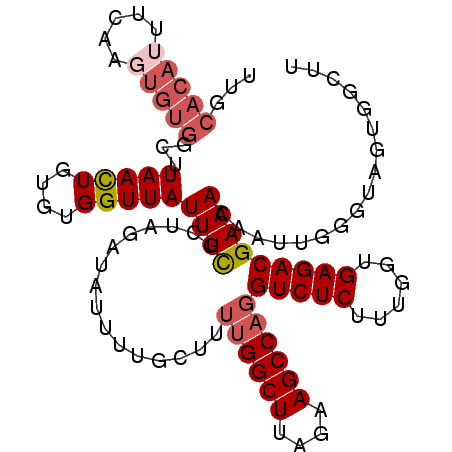
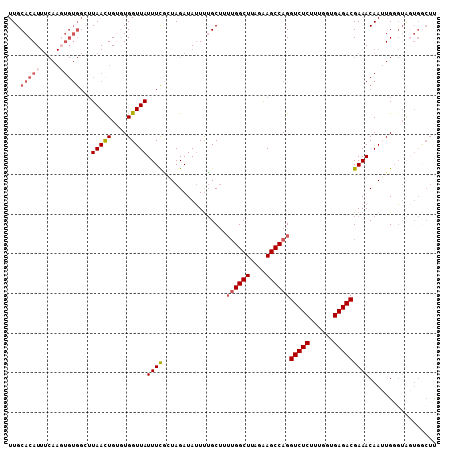
| Location | 1,008,432 – 1,008,527 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -20.64 |
| Consensus MFE | -15.28 |
| Energy contribution | -15.68 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.975719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1008432 95 - 22407834 AAGCCAAUAACCACUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAAGCAAAUUAUCUAGUGAAAUAACCACACAGUUA-------------AUGUGCAA ..((((.((((...((((((.(((((......))))).(((((....))))).))))))........(((.......)))...))))-------------.)).)).. ( -22.80) >DroSec_CAF1 120180 108 - 1 AAGCCACCACCCAAUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAAGCAGAAUAUCUACCGAAAUAACCACACAGUUAAGCCACACUUGAAAUGUGCAA ..............((((((((((((......))))).(((((....))))).................))))))).((((...(((((.....)))))..))))... ( -22.80) >DroSim_CAF1 120754 108 - 1 AAGCCACCACCCAAUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAUGCAAAAUAUCUACCAAAAUAACCACACAAUUAAGCCACACUUGAAAUGUGCAA ..((.................(((((......))))).(((((....)))))...))......................((((.(((((.....)))))..))))... ( -18.00) >DroEre_CAF1 125310 97 - 1 A-------UCACACUUGUUUCGUCUCAGCGCAGAGACCUGGCUCC-AAGCCCUUAGCCAAAUAUCUAUGGAAAUAACCA--CAGUUAAGCCACA-UUGAAAUGUGCAA .-------...(((..((((((((((......))))).(((((..-....((.(((........))).))...((((..--..)))))))))..-..))))))))... ( -19.50) >DroYak_CAF1 127308 106 - 1 GAGUCACUACACAAUUGUUUCGUCUCAGCAAAGAGACCUGGCUCCUAAGCCAAUAGCAAAAUAUCUAGCGAAAUAACCA--CAGUUAAUCCACACUUGAAAUGUGCAA .........((((.((((((((((((......))))).(((((....))))).................))))))).((--.(((........)))))...))))... ( -20.10) >consensus AAGCCACCACCCAAUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAAGCAAAAUAUCUACCGAAAUAACCACACAGUUAAGCCACACUUGAAAUGUGCAA ..............((((((((((((......))))).(((((....))))).................)))))))................................ (-15.28 = -15.68 + 0.40)

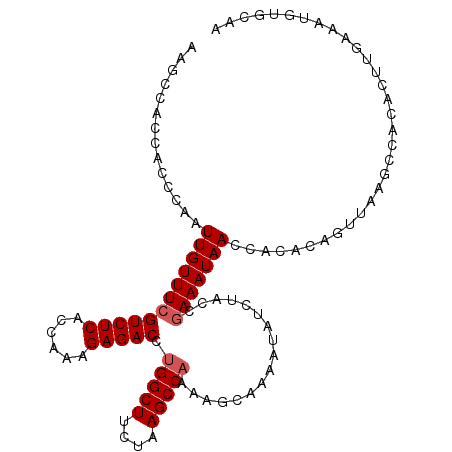
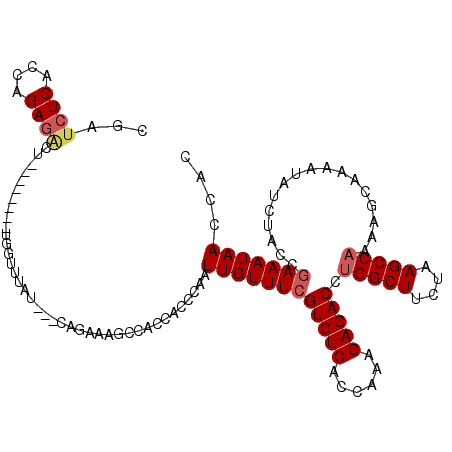
| Location | 1,008,447 – 1,008,556 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.23 |
| Mean single sequence MFE | -24.98 |
| Consensus MFE | -16.52 |
| Energy contribution | -17.20 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.992194 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 1008447 109 - 22407834 CGUUCUCACCAGAGACU--------UGGUUUUU---AAGAAAGCCAAUAACCACUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAAGCAAAUUAUCUAGUGAAAUAACCAC .....((((.(((...(--------(((((((.---...)))))))).......((((((.(((((......))))).(((((....))))).))))))....))).))))......... ( -27.80) >DroSec_CAF1 120208 109 - 1 CGGUCUCACCAGAGACU--------UGGUUUCU---CAGAAAGCCACCACCCAAUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAAGCAGAAUAUCUACCGAAAUAACCAC .((((((....))))))--------((((((..---....))))))........((((((((((((......))))).(((((....))))).................))))))).... ( -28.90) >DroSim_CAF1 120782 109 - 1 CGAUCUCACCAGAGACU--------UGGUUUUU---CAGAAAGCCACCACCCAAUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAUGCAAAAUAUCUACCAAAAUAACCAC ...((((....))))..--------(((((((.---...)))))))........((((...(((((......))))).(((((....)))))...))))..................... ( -22.10) >DroEre_CAF1 125336 103 - 1 CCAACUCACCAGAGACA--------GCGUUUAUAAUCGGAA-------UCACACUUGUUUCGUCUCAGCGCAGAGACCUGGCUCC-AAGCCCUUAGCCAAAUAUCUAUGGAAAUAACCA- (((.(((....)))...--------(((((......(((((-------(.......))))))....)))))(((....(((((..-........)))))....))).))).........- ( -19.80) >DroYak_CAF1 127335 119 - 1 CGAUCUCAGCAGAUGCGUCUCACAUGCGUUUAAAAUCCGAGAGUCACUACACAAUUGUUUCGUCUCAGCAAAGAGACCUGGCUCCUAAGCCAAUAGCAAAAUAUCUAGCGAAAUAACCA- .((((((.(.((((((((.....)))))))).....).)))).)).........((((((((((((......))))).(((((....))))).................)))))))...- ( -26.30) >consensus CGAUCUCACCAGAGACU________UGGUUUAU___CAGAAAGCCACCACCCAAUUGUUUCGUCUCACCAAAGAGACCUGGCUUCUAAGCCAAAAGCAAAAUAUCUACCGAAAUAACCAC ...((((....)))).......................................((((((((((((......))))).(((((....))))).................))))))).... (-16.52 = -17.20 + 0.68)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:33:13 2006