
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
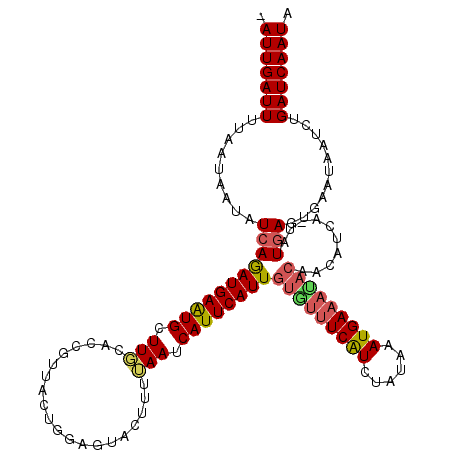
| Location | 9,634,955 – 9,635,073 |
| Length | 118 |
| Max. P | 0.986525 |

| Location | 9,634,955 – 9,635,073 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -27.14 |
| Consensus MFE | -16.56 |
| Energy contribution | -16.96 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.41 |
| Structure conservation index | 0.61 |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.986525 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
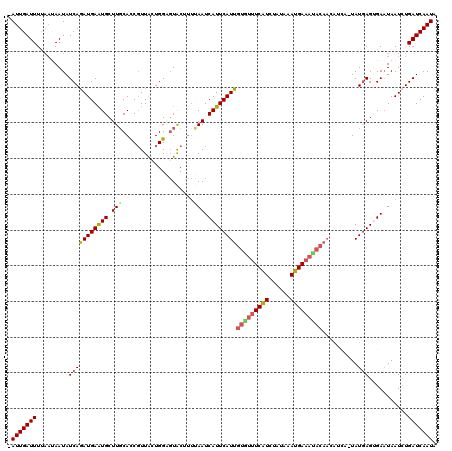
>2L_DroMel_CAF1 9634955 118 + 22407834 -AUUGAUUUUCAAAAUAUCAAAUGAAUGCUUGCACCGUUACUGGAGUACAUUUAAUCAUUCAUUGUGUUUCAUCUAUAAAUGAAAUACAACAUUA-UAUGAGUGAAUAAUCUGAUCAAUA -(((((((..............((((((..(((.(((....))).))))))))).((((((((((((((((((......))))))))))......-.)))))))).......))))))). ( -26.51) >DroSec_CAF1 35557 118 + 1 -AUUGAUUUUAAUAAUAUCAGAUGAAUGCUUGCACCGUUAGUGGAGUACUUUUAAUCAUUCAUUGUGUUUCAUUUAAAAAUGAAACACAACAUUA-UAUGAGUGAAUAAUCUGAUCAAUA -...............(((((((...(((((.(((.....)))))))).......(((((((((((((((((((....)))))))))))......-.))))))))...)))))))..... ( -30.21) >DroSim_CAF1 34846 108 + 1 -AUUGAUUUUAAUAAUAUCAGAUGAAUGCUUGCACCGUUACUGGAGUACUUUUAAUCAUUCAUUGUGUUUCAUUUAAAAAUGAC----------A-UAUGAGUGAAUAAUCUGAUCAAUA -...............(((((((.......(((.(((....))).))).......((((((((.(((..(((((....))))))----------)-)))))))))...)))))))..... ( -20.90) >DroEre_CAF1 37721 116 + 1 AAUUGAUUUCACCAAUAUCAGAUGAAUGCUUGCAUCCUUGCUGGUUUGC----AAUCAUUCAUUGUAUUUCGUCUAUAAAUGAAAUACAACAUCUUUAUGAGAGAUUAAUAUGAUCAAUA .(((((((((..........((((((((.(((((.((.....))..)))----)).))))))))(((((((((......))))))))).............))))))))).......... ( -32.20) >DroYak_CAF1 37350 119 + 1 AAUUGAUUUCAAUAAUAUCAGAUGAGUGCUUUCAUCGUUAUUGGUGUACUUUAAAUCAUUCAUUGUAUUUCAUCUAUAAAUGAAAUACAACAUCU-UAUAAGCGAAUAAUCUGAUCAAUA .((((....))))...(((((((...(((((.(((((....)))))................(((((((((((......))))))))))).....-...)))))....)))))))..... ( -25.90) >consensus _AUUGAUUUUAAUAAUAUCAGAUGAAUGCUUGCACCGUUACUGGAGUACUUUUAAUCAUUCAUUGUGUUUCAUCUAUAAAUGAAAUACAACAUCA_UAUGAGUGAAUAAUCUGAUCAAUA .(((((((.........(((((((((((.(((....................))).))))))))(((((((((......)))))))))..........)))...........))))))). (-16.56 = -16.96 + 0.40)

| Location | 9,634,955 – 9,635,073 |
|---|---|
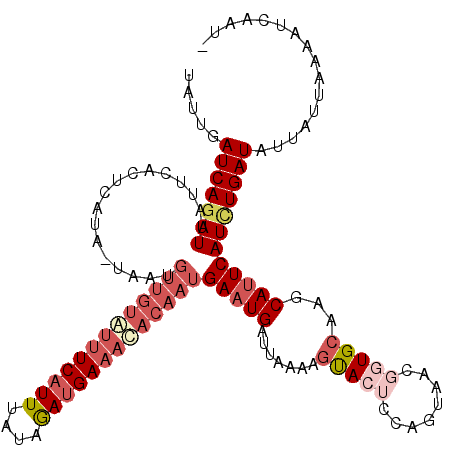
| Length | 118 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.38 |
| Mean single sequence MFE | -25.34 |
| Consensus MFE | -17.36 |
| Energy contribution | -20.08 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.986197 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
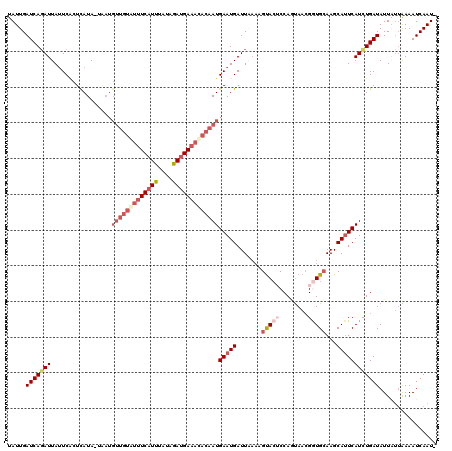
>2L_DroMel_CAF1 9634955 118 - 22407834 UAUUGAUCAGAUUAUUCACUCAUA-UAAUGUUGUAUUUCAUUUAUAGAUGAAACACAAUGAAUGAUUAAAUGUACUCCAGUAACGGUGCAAGCAUUCAUUUGAUAUUUUGAAAAUCAAU- .((((((((((.((((........-....(((((.(((((((....))))))).)))))((((((.....((((((........)))))).....)))))))))).)))))...)))))- ( -24.50) >DroSec_CAF1 35557 118 - 1 UAUUGAUCAGAUUAUUCACUCAUA-UAAUGUUGUGUUUCAUUUUUAAAUGAAACACAAUGAAUGAUUAAAAGUACUCCACUAACGGUGCAAGCAUUCAUCUGAUAUUAUUAAAAUCAAU- .....(((((((............-....(((((((((((((....)))))))))))))(((((.......(((((........)))))...))))))))))))...............- ( -28.50) >DroSim_CAF1 34846 108 - 1 UAUUGAUCAGAUUAUUCACUCAUA-U----------GUCAUUUUUAAAUGAAACACAAUGAAUGAUUAAAAGUACUCCAGUAACGGUGCAAGCAUUCAUCUGAUAUUAUUAAAAUCAAU- .....(((((((((.(((.((((.-.----------.(((((....)))))......)))).))).)))..(((((........))))).........))))))...............- ( -17.70) >DroEre_CAF1 37721 116 - 1 UAUUGAUCAUAUUAAUCUCUCAUAAAGAUGUUGUAUUUCAUUUAUAGACGAAAUACAAUGAAUGAUU----GCAAACCAGCAAGGAUGCAAGCAUUCAUCUGAUAUUGGUGAAAUCAAUU .((((((..(((((((.((......((((((((((((((.((....)).))))))))))(((((.((----(((..((.....)).))))).))))))))))).)))))))..)))))). ( -28.70) >DroYak_CAF1 37350 119 - 1 UAUUGAUCAGAUUAUUCGCUUAUA-AGAUGUUGUAUUUCAUUUAUAGAUGAAAUACAAUGAAUGAUUUAAAGUACACCAAUAACGAUGAAAGCACUCAUCUGAUAUUAUUGAAAUCAAUU .((((((.(((((((((.((....-))..(((((((((((((....)))))))))))))))))))))).........((((((.(((((......))))).....))))))..)))))). ( -27.30) >consensus UAUUGAUCAGAUUAUUCACUCAUA_UAAUGUUGUAUUUCAUUUAUAGAUGAAACACAAUGAAUGAUUAAAAGUACUCCAGUAACGGUGCAAGCAUUCAUCUGAUAUUAUUAAAAUCAAU_ .....(((((((.................(((((((((((((....)))))))))))))(((((.......(((((........)))))...))))))))))))................ (-17.36 = -20.08 + 2.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:53 2006