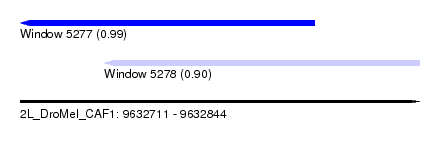
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,632,711 – 9,632,844 |
| Length | 133 |
| Max. P | 0.989686 |

| Location | 9,632,711 – 9,632,809 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.25 |
| Mean single sequence MFE | -39.88 |
| Consensus MFE | -23.47 |
| Energy contribution | -24.79 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.59 |
| SVM decision value | 2.18 |
| SVM RNA-class probability | 0.989686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
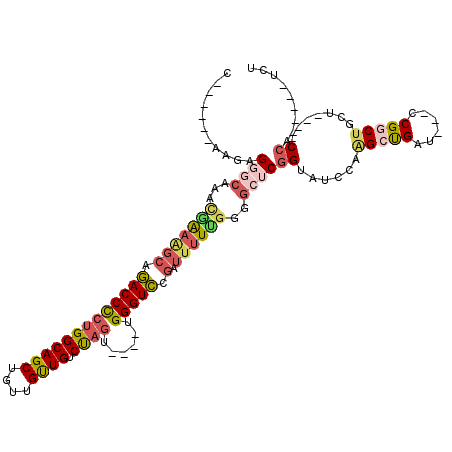
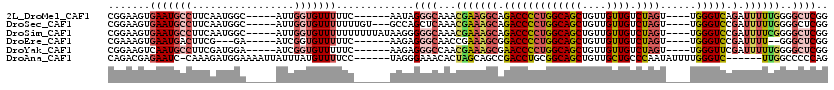
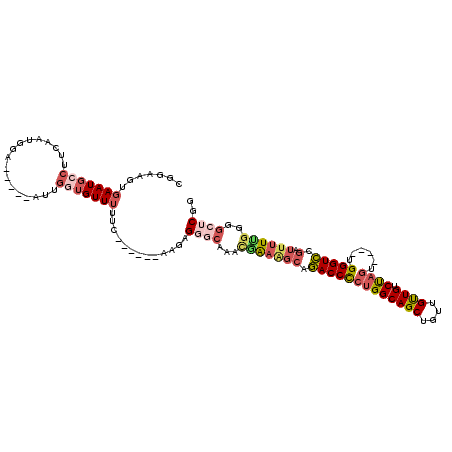
>2L_DroMel_CAF1 9632711 98 - 22407834 C------AAUAGGGCAAACGAAGGCAGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCAGAUUUUUGGGGCUCGGUAUCCAAGCUGAU---CCGGCUGCU----CCA-----UCU .------....(((((..((..(((.(((((((((((((....)))).)))).----.)))))......(((((........))))))))...---.))..))).----)).-----... ( -33.10) >DroSec_CAF1 33367 101 - 1 UUGU---GCCAGCUCAAACGAAAGCAGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCCGAUUUUUGGGGCUCGGUAUCCAAGCUGAU---CCGCCUGCU----CCA-----UCU ....---....(((........))).(((((((((((((....)))).)))).----.))))).......((((((.(((.(((......)))---)))...)))----)))-----... ( -31.70) >DroSim_CAF1 32574 104 - 1 UUUUUAUAAGGGGGCAAACGAAAGCAGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCCGAUUUUCGGGGCUCGGUAUACAAGCUGAU---CCGCCAGCU----CCA-----UCU ..........(((((....((((((.(((((((((((((....)))).)))).----.))))).).)))))((((.(((((......))))).---)).)).)))----)).-----... ( -39.70) >DroEre_CAF1 35507 96 - 1 C------AAGAGGGCAACCGAAAGCGGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCCGAUUUU--GGGCUCGGUAUCCAAGCUGAU---CCGGCUGCG----CCA-----UCU .------.(((.(((..((((((.(((((((((((((((....)))).)))).----.))))))).))))--))((.(((.(((......)))---)))))...)----)).-----))) ( -43.60) >DroYak_CAF1 35026 98 - 1 C------AAGAGGGCCAACGAAAGCGAACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUUCGAUUUUUGGGGCUCGGUAUCCAAGCCGAU---CCGGCUGCU----CCA-----UCU .------..(((((((..(((((((((((((((((((((....)))).)))).----.))))))).))))))(((.(((((......))))))---)))))).))----)..-----... ( -42.90) >DroAna_CAF1 44508 107 - 1 C------UAGGGAAACACUAGCAGCCGACCUGCGGCAGCUGUUGCUGCCCAAUAUUUUGGGUC------UUGGCCCCCAGUCUCC-GGCUGGUGUGCCGGCUGGUUUUUCCACUGAUUUC .------((((((((.((((((....((((...((((((....))))))(((....)))))))------.(((((.(((((....-.))))).).)))))))))).))))).)))..... ( -48.30) >consensus C______AAGAGGGCAAACGAAAGCAGACCCCUGGCAGCUGUUGUUGUCUAGU____UGGGUCCGAUUUUUGGGGCUCGGUAUCCAAGCUGAU___CCGGCUGCU____CCA_____UCU ...........((((...(((((((.(((((((((((((....)))).))))......))))).).))))))..))))((......(((((......))))).......))......... (-23.47 = -24.79 + 1.31)

| Location | 9,632,739 – 9,632,844 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.61 |
| Mean single sequence MFE | -37.48 |
| Consensus MFE | -19.24 |
| Energy contribution | -20.96 |
| Covariance contribution | 1.73 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.51 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897787 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9632739 105 - 22407834 CGGAAGUGAAUGCCUUCAAUGGC-----AUUGGUGUUUUUC------AAUAGGGCAAACGAAGGCAGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCAGAUUUUUGGGGCUCGG .((((((.((((((......)))-----))).)...)))))------....((((...(((((((.(((((((((((((....)))).)))).----.))))).).))))))..)))).. ( -35.80) >DroSec_CAF1 33395 108 - 1 CGGAAGUGAAUGCCUUCAAUGGC-----AUUGGUGUUUUUUUGU---GCCAGCUCAAACGAAAGCAGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCCGAUUUUUGGGGCUCGG (....)..((((((......)))-----)))((..(......).---.))(((((...(((((((.(((((((((((((....)))).)))).----.))))).).)))))))))))... ( -36.10) >DroSim_CAF1 32602 111 - 1 CGGAAGUGAAUGCCUUCAAUGGC-----AUUGGUGUUUUUUUUUUAUAAGGGGGCAAACGAAAGCAGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCCGAUUUUCGGGGCUCGG .(((..(.((((((......)))-----))).)..))).............((((...(((((((.(((((((((((((....)))).)))).----.))))).).))))))..)))).. ( -37.50) >DroEre_CAF1 35535 100 - 1 CGAAAGUGAAUGACUUCG---GA-----AUCGGUGUUUUUC------AAGAGGGCAACCGAAAGCGGACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUCCGAUUUU--GGGCUCGG (....)((((.(((.(((---..-----..))).))).)))------)...((((..((((((.(((((((((((((((....)))).)))).----.))))))).))))--)))))).. ( -39.60) >DroYak_CAF1 35054 105 - 1 CGGAAGUCAAUGCCUUCGAUGGA-----AUCGGUGUUUUUC------AAGAGGGCCAACGAAAGCGAACCCCUGGCAGCUGUUGUUGUCUAGU----UGGGUUCGAUUUUUGGGGCUCGG ((..((((...((((((..((((-----(........))))------).))))))...(((((((((((((((((((((....)))).)))).----.))))))).)))))).)))))). ( -39.40) >DroAna_CAF1 44547 107 - 1 CAGACGAGAAUC-CAAAGAUGGAAAAUUAUUUAUGUUUUCC------UAGGGAAACACUAGCAGCCGACCUGCGGCAGCUGUUGCUGCCCAAUAUUUUGGGUC------UUGGCCCCCAG ....(((((.((-(((((..(((((((.......)))))))------((((....).)))((((.....))))((((((....)))))).....)))))))))------)))........ ( -36.50) >consensus CGGAAGUGAAUGCCUUCAAUGGA_____AUUGGUGUUUUUC______AAGAGGGCAAACGAAAGCAGACCCCUGGCAGCUGUUGUUGUCUAGU____UGGGUCCGAUUUUUGGGGCUCGG .......(((((((.................))))))).............((((...(((((((.(((((((((((((....)))).))))......))))).).))))))..)))).. (-19.24 = -20.96 + 1.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:50 2006