
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,626,880 – 9,626,988 |
| Length | 108 |
| Max. P | 0.823977 |

| Location | 9,626,880 – 9,626,988 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.11 |
| Mean single sequence MFE | -36.35 |
| Consensus MFE | -19.42 |
| Energy contribution | -21.87 |
| Covariance contribution | 2.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
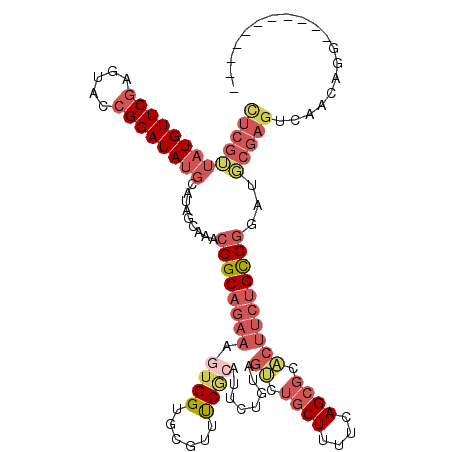
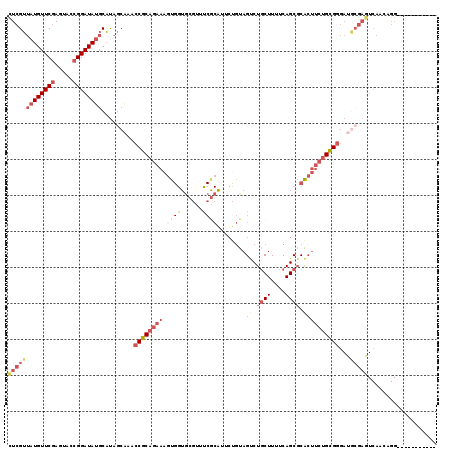
>2L_DroMel_CAF1 9626880 108 + 22407834 CUCGUUAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUUCGCAUUCUGUAGUCUGCUUUUCAGCGCACUUCUGCGGGAUGCGAGUCAACAGG----------- (((((.(((((((.....)))))))))...(((..(((((((....((((((((..(((..(....)..))).....)))))))))))))))..))))))........----------- ( -38.90) >DroSec_CAF1 27522 108 + 1 CUCGUUAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUCCGCAUUCUGUAGUCUGCUUUUCAGCGCACUUCUGCGGGAUGCGAGUCAACAGG----------- (((((.(((((((.....)))))))))...(((..(((((((....((((((((..(((..(....)..))).....)))))))))))))))..))))))........----------- ( -38.90) >DroSim_CAF1 26733 108 + 1 CUCGUUAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUCCGCAUUCUGUAGUCUGCUUUUCAGCUCACUUCUGCGGGAUGCGAGUCAACAGG----------- (((((.(((((((.....)))))))))...(((..((((((((.((((.((.....(((..(....)..)))......))))))))))))))..))))))........----------- ( -37.30) >DroEre_CAF1 29540 96 + 1 CUCGUUAUGUUCGAGUAACGGAUAUGCACAGCGAACCGCAGAAAGUGGUGCGUAUCGCAGACUGUAGUCUGCUUUUCAGCGCACUUCUGCGGC-----------------------AGC ...((((((((((.....)))))))).)).((...(((((((....(((((((...((((((....))))))......)))))))))))))).-----------------------.)) ( -38.80) >DroYak_CAF1 28925 102 + 1 CUCGUUAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGU-----------------AUAGCCUGCUUUUCAGCGCACUUCUGCGGGAUGCGAGUCAACAGGCAGCGGUGAGC ((((.......))))(((((..........(((..((((((((.((-----------------...(((((.....))).))))))))))))..)))..(((....)))..)))))... ( -33.90) >DroAna_CAF1 37126 103 + 1 UUCGAUUUGUUCAAGUACCGGAUAUCCUUUACCGACCGCAA----UGGUAG--UUCACGAACCGUAGCCUACUUUUCAGCGCGGCUCGGUGAGACUCGAGUCAAGUGG----------C ..(.((((((((.(((...((....))((((((((((((.(----((((.(--....)..))))).((..........)))))).))))))))))).))).))))).)----------. ( -30.30) >consensus CUCGUUAUGUUCGAGUACCGGAUAUGCAUAGCAAACCGCAGAAAGUGGUGCGUUUCGCAUUCUGUAGUCUGCUUUUCAGCGCACUUCUGCGGGAUGCGAGUCAACAGG___________ (((((((((((((.....)))))))).........((((((((.((((......))))........((.((((....)))).))))))))))...)))))................... (-19.42 = -21.87 + 2.45)

| Location | 9,626,880 – 9,626,988 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.11 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -18.78 |
| Energy contribution | -20.63 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755285 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
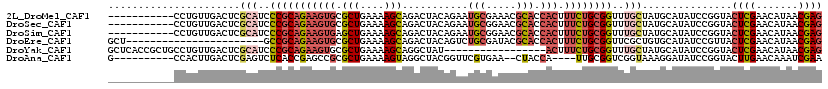
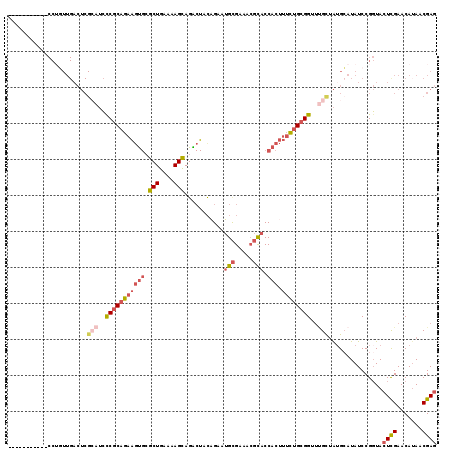
>2L_DroMel_CAF1 9626880 108 - 22407834 -----------CCUGUUGACUCGCAUCCCGCAGAAGUGCGCUGAAAAGCAGACUACAGAAUGCGAAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUAACGAG -----------((.(.((....(((..(((((((((((.(((....)))...........((((...)))).))).))))))))..)))....))...).))..((((.......)))) ( -33.30) >DroSec_CAF1 27522 108 - 1 -----------CCUGUUGACUCGCAUCCCGCAGAAGUGCGCUGAAAAGCAGACUACAGAAUGCGGAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUAACGAG -----------((.(.((....(((..(((((((((((.(((....)))...........((((...)))).))).))))))))..)))....))...).))..((((.......)))) ( -33.30) >DroSim_CAF1 26733 108 - 1 -----------CCUGUUGACUCGCAUCCCGCAGAAGUGAGCUGAAAAGCAGACUACAGAAUGCGGAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUAACGAG -----------((.(.((....(((..(((((((((((.(((....)))...........((((...)))).))).))))))))..)))....))...).))..((((.......)))) ( -33.90) >DroEre_CAF1 29540 96 - 1 GCU-----------------------GCCGCAGAAGUGCGCUGAAAAGCAGACUACAGUCUGCGAUACGCACCACUUUCUGCGGUUCGCUGUGCAUAUCCGUUACUCGAACAUAACGAG ((.-----------------------((((((((((((((.......((((((....))))))....)))))....)))))))))..))..........(((((........))))).. ( -35.20) >DroYak_CAF1 28925 102 - 1 GCUCACCGCUGCCUGUUGACUCGCAUCCCGCAGAAGUGCGCUGAAAAGCAGGCUAU-----------------ACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUAACGAG ((.....))((((.(.((....(((..(((((((((((((((....)))..))...-----------------)).))))))))..)))....))...).))))((((.......)))) ( -30.90) >DroAna_CAF1 37126 103 - 1 G----------CCACUUGACUCGAGUCUCACCGAGCCGCGCUGAAAAGUAGGCUACGGUUCGUGAA--CUACCA----UUGCGGUCGGUAAAGGAUAUCCGGUACUUGAACAAAUCGAA .----------.........((((((..((((((.(((((.((....((((..((((...))))..--))))))----.))))))))))...((....)))..)))))).......... ( -30.80) >consensus ___________CCUGUUGACUCGCAUCCCGCAGAAGUGCGCUGAAAAGCAGACUACAGAAUGCGAAACGCACCACUUUCUGCGGUUUGCUAUGCAUAUCCGGUACUCGAACAUAACGAG ......................(((..(((((((((((.(((....)))...........(((.....))).))).))))))))..)))...............((((.......)))) (-18.78 = -20.63 + 1.85)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:47 2006