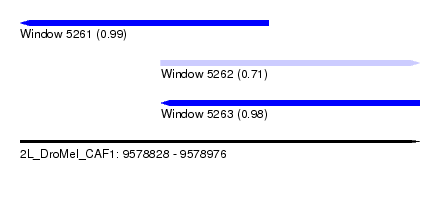
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,578,828 – 9,578,976 |
| Length | 148 |
| Max. P | 0.994103 |

| Location | 9,578,828 – 9,578,920 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 64.83 |
| Mean single sequence MFE | -24.82 |
| Consensus MFE | -16.27 |
| Energy contribution | -16.40 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.66 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9578828 92 - 22407834 UUCAACCGGCAGCUCAGGCAGCUGAUGAUGAUAACAAUAUGACAAAUUCAAAUCCGGAGGUUCCAGAUGUCGG--AGGCAAAU---------------CAGAAGGAUCA (((..(((((((((.....)))))...............(((.....)))...)))).(.((((.......))--)).)....---------------..)))...... ( -17.40) >DroSec_CAF1 6908 69 - 1 -----CCGGCAGCUCAGGCUGCUGAGGACAAUA------------------AUCCGGAGGUUCCAAAUGCCAA--AAGCAAAU---------------CAGGAGGAUCA -----.(((((((....))))))).(((.....------------------.))).((..((((...(((...--..)))...---------------..))))..)). ( -20.90) >DroSim_CAF1 6565 69 - 1 -----CCGGCAGCUCAGGCUGCUGAGGAUAAUA------------------AUCCGGAGGUUCCAGAUGCCAA--AAGCAAAU---------------CAGGAGGAUCA -----.(((((((....))))))).((((....------------------)))).((..((((.(((((...--..)))..)---------------).))))..)). ( -23.70) >DroEre_CAF1 7020 106 - 1 UUCAUCCGGCAGCUCAGGCUGCCGAUGAAAAUAACAAUGUGCAAAAUCAGAAUCCGCAGGCUCCAGAUGCCGAUGAUGCAAAUGGGCGGGUUGAAGUGCAGGAGGA--- ((((((.((((((....))))))))))))........(.((((...(((..((((((.....(((..(((.......)))..))))))))))))..)))).)....--- ( -37.30) >consensus _____CCGGCAGCUCAGGCUGCUGAGGAUAAUA__________________AUCCGGAGGUUCCAGAUGCCAA__AAGCAAAU_______________CAGGAGGAUCA ......(((((((....)))))))............................(((((.....))...(((.......))).......................)))... (-16.27 = -16.40 + 0.13)


| Location | 9,578,880 – 9,578,976 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 70.56 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -12.00 |
| Energy contribution | -12.25 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.47 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711211 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
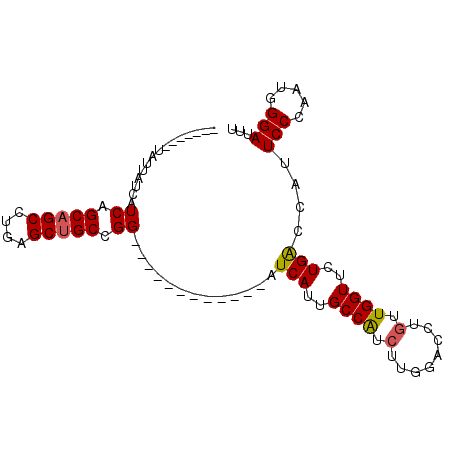
>2L_DroMel_CAF1 9578880 96 + 22407834 AUAUUGUUAUCAUCAUCAGCUGCCUGAGCUGCCGGUUGAAGAUUCCGAUCAUUGCCGUCUUCGACCUGUUGGUUCUGACCAUUCCGACUGGGAUUU ...........(((..(((((.....)))))((((((((((((..(((...)))..)))))))))).(((((...........))))).))))).. ( -25.50) >DroSec_CAF1 6949 77 + 1 -------UAUUGUCCUCAGCAGCCUGAGCUGCCGG------------AUCAUUGCCAUCUUGGACCUGUUGGUUCUGACCAUUCCCAAUGGGAUUU -------....((((.(((((((....))))).((------------.(((..((((.(........).))))..)))))........)))))).. ( -21.80) >DroSim_CAF1 6606 77 + 1 -------UAUUAUCCUCAGCAGCCUGAGCUGCCGG------------AUCAUUGCCAUCUUGGACCUGUUGGUUCUGACCAUUCCCAAUGGGAUUU -------....((((.(((((((....))))).((------------.(((..((((.(........).))))..)))))........)))))).. ( -21.50) >DroEre_CAF1 7086 84 + 1 ACAUUGUUAUUUUCAUCGGCAGCCUGAGCUGCCGGAUGAAGAUUCCGGUCAUUGCCGCCUUGAGCCUCUUGGUGCUGGCCAU------------UU .....(..(((((((((((((((....)))))).)))))))))..)(((((..((.(((..(.....)..))))))))))..------------.. ( -33.70) >consensus _______UAUUAUCAUCAGCAGCCUGAGCUGCCGG____________AUCAUUGCCAUCUUGGACCUGUUGGUUCUGACCAUUCCCAAUGGGAUUU ...............((.(((((....))))).)).............(((..((((.(........).))))..)))....(((.....)))... (-12.00 = -12.25 + 0.25)


| Location | 9,578,880 – 9,578,976 |
|---|---|
| Length | 96 |
| Sequences | 4 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 70.56 |
| Mean single sequence MFE | -23.85 |
| Consensus MFE | -16.30 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978859 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9578880 96 - 22407834 AAAUCCCAGUCGGAAUGGUCAGAACCAACAGGUCGAAGACGGCAAUGAUCGGAAUCUUCAACCGGCAGCUCAGGCAGCUGAUGAUGAUAACAAUAU .....((.((((...((((....))))....((((....))))..)))).)).(((.(((..((((.((....)).)))).))).)))........ ( -21.00) >DroSec_CAF1 6949 77 - 1 AAAUCCCAUUGGGAAUGGUCAGAACCAACAGGUCCAAGAUGGCAAUGAU------------CCGGCAGCUCAGGCUGCUGAGGACAAUA------- ...((((...)))).((((....))))....((((..(((.......))------------)(((((((....))))))).))))....------- ( -25.00) >DroSim_CAF1 6606 77 - 1 AAAUCCCAUUGGGAAUGGUCAGAACCAACAGGUCCAAGAUGGCAAUGAU------------CCGGCAGCUCAGGCUGCUGAGGAUAAUA------- ...((((...)))).((((....))))....((((..(((.......))------------)(((((((....))))))).))))....------- ( -23.10) >DroEre_CAF1 7086 84 - 1 AA------------AUGGCCAGCACCAAGAGGCUCAAGGCGGCAAUGACCGGAAUCUUCAUCCGGCAGCUCAGGCUGCCGAUGAAAAUAACAAUGU ..------------...((((((........)))...)))((......))(..((.((((((.((((((....)))))))))))).))..)..... ( -26.30) >consensus AAAUCCCAUUGGGAAUGGUCAGAACCAACAGGUCCAAGACGGCAAUGAU____________CCGGCAGCUCAGGCUGCUGAGGAUAAUA_______ ...(((.....))).(((......)))....(((......)))...................(((((((....)))))))................ (-16.30 = -16.18 + -0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:35 2006