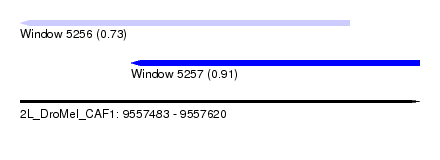
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,557,483 – 9,557,620 |
| Length | 137 |
| Max. P | 0.908073 |

| Location | 9,557,483 – 9,557,596 |
|---|---|
| Length | 113 |
| Sequences | 4 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 76.31 |
| Mean single sequence MFE | -33.55 |
| Consensus MFE | -21.41 |
| Energy contribution | -21.22 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726406 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9557483 113 - 22407834 CAUGGCGUAUACGCGAUUUACCACGACUCUUGAUUCAAAUUGAGGAAUGCUGAAACAUUGCGUAUACGCUACUUAAGUUAGCCAGUCUGUUGAUUCCCUUAAA-GUGCGUGUAG- (((((((((((((((((....((((..(((..((....))..)))..)).))....)))))))))))))(((((((((((((......)))))....))).))-)))))))...- ( -29.20) >DroSim_CAF1 9231 113 - 1 CAUGGCGUAUACGCGAUUUACCGCGACUCUUGAUUCAAAUUGAGGAAUGCUGCAACAUUGCGUAUACGCUACUUAAGUUAGCCAGUCUGUUGAUUCCCUUAUA-GUGCUUGUAG- ..(((((((((((((((((.(.(((..(((..((....))..)))..))).).)).)))))))))))))))..(((((.((..((((....))))..))....-..)))))...- ( -31.00) >DroEre_CAF1 8400 109 - 1 CGUGGCGUAUACGCGAUUUGGCCCGACUGUGGAUUCGCAUUGAGGAAUGCUGCAACAUGGCGUAUGCGCUAGUUGGCUUAGUCAGUCUGCUGAG-----CAGUCAUGCCUUU-UC ..((((((((((((.((...(((((....)))....(((((....))))).))...)).))))))))))))....(((((((......))))))-----)............-.. ( -37.00) >DroYak_CAF1 7443 104 - 1 CAUGGCGUAUACGCAA-----------UGUUAAUUCGCAUUGAGGAAUGCUGCAACAUGGCGUAUACGCUACUUAACUUGGCCCGUCUGUUGAUUUUCAUAGACAUGCCUUUGGC ..((((((((((((.(-----------((((.....(((((....)))))...))))).))))))))))))........(((..((((((.(.....)))))))..)))...... ( -37.00) >consensus CAUGGCGUAUACGCGAUUUACC_CGACUCUUGAUUCAAAUUGAGGAAUGCUGCAACAUGGCGUAUACGCUACUUAACUUAGCCAGUCUGUUGAUUCCCUUAGA_AUGCCUGUAG_ ..((((((((((((.............(((((((....))))))).(((......))).)))))))))))).....((((((......))))))..................... (-21.41 = -21.22 + -0.19)

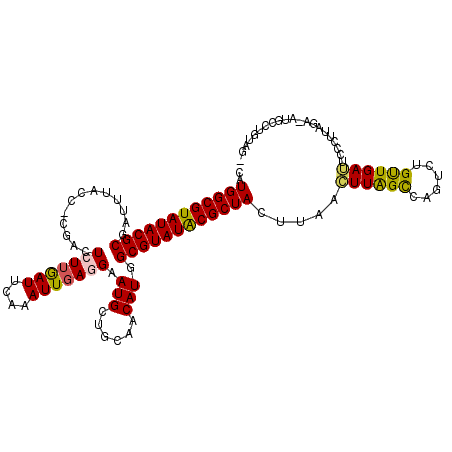
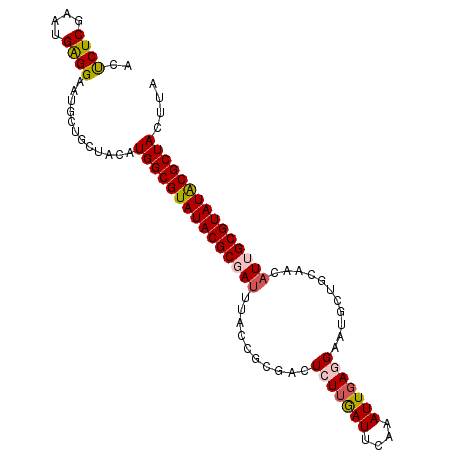
| Location | 9,557,521 – 9,557,620 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 86.57 |
| Mean single sequence MFE | -31.10 |
| Consensus MFE | -22.42 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908073 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9557521 99 - 22407834 ACUCUCGAAUGAGGAAUGCUGCUACAUGGCGUAUACGCGAUUUACCACGACUCUUGAUUCAAAUUGAGGAAUGCUGAAACAUUGCGUAUACGCUACUUA .(((......))).............(((((((((((((((....((((..(((..((....))..)))..)).))....))))))))))))))).... ( -28.40) >DroSec_CAF1 7925 99 - 1 ACUCUCGAAUGAGGAAUGCUGCUACAUGGCGUAUACGCGAUUUACCGCGACUCUUGAUUCAAAUUGAGGAAUGCUGCAACAUUGCGUAUACGCUACUUA .(((......))).............(((((((((((((((((.(.(((..(((..((....))..)))..))).).)).))))))))))))))).... ( -30.80) >DroSim_CAF1 9269 99 - 1 ACUCUCGAAUGAGGAAUGCUGCUACAUGGCGUAUACGCGAUUUACCGCGACUCUUGAUUCAAAUUGAGGAAUGCUGCAACAUUGCGUAUACGCUACUUA .(((......))).............(((((((((((((((((.(.(((..(((..((....))..)))..))).).)).))))))))))))))).... ( -30.80) >DroEre_CAF1 8434 99 - 1 CCCCUCGAUUGAGGAAUGCUGCAACGUGGCGUAUACGCGAUUUGGCCCGACUGUGGAUUCGCAUUGAGGAAUGCUGCAACAUGGCGUAUGCGCUAGUUG ..((((....)))).......((((.((((((((((((.((...(((((....)))....(((((....))))).))...)).)))))))))))))))) ( -36.20) >DroYak_CAF1 7483 88 - 1 ACUCUCGAAUGGGGAAUGCUGCAACAUGGCGUAUACGCAA-----------UGUUAAUUCGCAUUGAGGAAUGCUGCAACAUGGCGUAUACGCUACUUA ..((((.....))))...........((((((((((((.(-----------((((.....(((((....)))))...))))).)))))))))))).... ( -29.30) >consensus ACUCUCGAAUGAGGAAUGCUGCUACAUGGCGUAUACGCGAUUUACCGCGACUCUUGAUUCAAAUUGAGGAAUGCUGCAACAUUGCGUAUACGCUACUUA ..((((....))))............(((((((((((((((..........(((((((....)))))))...........))))))))))))))).... (-22.42 = -22.98 + 0.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:29 2006