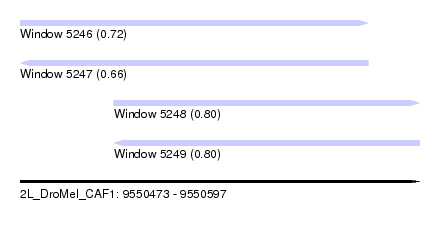
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,550,473 – 9,550,597 |
| Length | 124 |
| Max. P | 0.798837 |

| Location | 9,550,473 – 9,550,581 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -22.85 |
| Energy contribution | -23.43 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719562 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9550473 108 + 22407834 -----------UCCACCUCGACGGGGGGCUCAACGAUCCAGCUUUCGCUGGCUCUUCUGGCCAACAAAGCCGGCGAGAACAUGAGGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUG- -----------........(((((((((((((..((.(((((....))))).))((((.(((.........))).))))..))).)))))))..((((......))))....)))....- ( -38.60) >DroVir_CAF1 3206 106 + 1 -------------CACCUCAACGGGCGGCUCCACUAUCCAGCUCUCGCUGGCGCGACGCGCCAACAGCGCCGGCGAGAACAUCAAGUCCUCCUCAUCCUCCUCUGGAUACCAGUCCAUG- -------------.........((((((......(((((((.((((((((((((............))))))))))))((.....))...............))))))))).))))...- ( -37.10) >DroGri_CAF1 3667 110 + 1 ---------GCUGCACCUCAACGGGCGGUUCAACUAUCCAACUUUCACUAGCGCGACGCGCCAACAACGCCGGCGAGAACAUCAAGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUG- ---------(((((.((.....))))))).....(((((...........((((...))))..........((.(((.((.....)).))).))..........)))))..........- ( -21.60) >DroWil_CAF1 5768 119 + 1 UAGUCAUUAGACGAACCUCCACUGGUGGCUCAACGAUCCAACUCUCGCUGGCUCGUCGCGCCAAAAGUGCCGGCGAGAACAUCAGAUCCUCUUCAUCUUCCUCGGGAUACCAAUCCAUU- .(((((((((...........)))))))))....((((....((((((((((.(............).))))))))))......))))................((((....))))...- ( -32.40) >DroMoj_CAF1 3366 106 + 1 -------------CACCUCAACGGGCGGCUCCACUAUCCAGCUUUCGCUGGCCCGUCGCGCCAACAGGGCCGGCGAGAACAUCAAGUCCUCCUCAUCCUCCUCGGGAUACCAGUCCAUG- -------------.........((((((............(.((((((((((((............)))))))))))).)..............(((((....))))).)).))))...- ( -34.60) >DroAna_CAF1 1886 109 + 1 -----------GCCACCUCAACUGGUGGUUCAACUAUCCAGCUUUCACUGGCCCGCCUGGCCAGCAGAGCUGGCGAGAACAUGAGGUCCUCCUCAUCCUCCUCCGGAUACCAAUCCAUGG -----------((((((......))))))........((((((((..((((((.....)))))).)))))))).(((...((((((....)))))).)))....((((....)))).... ( -44.20) >consensus _____________CACCUCAACGGGCGGCUCAACUAUCCAGCUUUCGCUGGCCCGUCGCGCCAACAGAGCCGGCGAGAACAUCAAGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUG_ ......................((((((..............((((((((((((............))))))))))))................((((......)))).)).)))).... (-22.85 = -23.43 + 0.59)

| Location | 9,550,473 – 9,550,581 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.38 |
| Mean single sequence MFE | -43.85 |
| Consensus MFE | -31.55 |
| Energy contribution | -32.05 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.655638 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9550473 108 - 22407834 -CAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACCUCAUGUUCUCGCCGGCUUUGUUGGCCAGAAGAGCCAGCGAAAGCUGGAUCGUUGAGCCCCCCGUCGAGGUGGA----------- -...((.(((((..((((.((((.((((((....)))))).))))..))))........)))))..((.(((((....))))).))......))((((.....)).)).----------- ( -43.70) >DroVir_CAF1 3206 106 - 1 -CAUGGACUGGUAUCCAGAGGAGGAUGAGGAGGACUUGAUGUUCUCGCCGGCGCUGUUGGCGCGUCGCGCCAGCGAGAGCUGGAUAGUGGAGCCGCCCGUUGAGGUG------------- -......(((((((((......))))...((((((.....)))))))))))((((...(((((..(((.(((((....)))))...)))..).))))......))))------------- ( -42.60) >DroGri_CAF1 3667 110 - 1 -CAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACUUGAUGUUCUCGCCGGCGUUGUUGGCGCGUCGCGCUAGUGAAAGUUGGAUAGUUGAACCGCCCGUUGAGGUGCAGC--------- -..(.(((((.(((((......)))))..((((((.....)))))).(((((.(..(((((((...)))))))..)..))))).))))).)...((((.....)).))...--------- ( -36.90) >DroWil_CAF1 5768 119 - 1 -AAUGGAUUGGUAUCCCGAGGAAGAUGAAGAGGAUCUGAUGUUCUCGCCGGCACUUUUGGCGCGACGAGCCAGCGAGAGUUGGAUCGUUGAGCCACCAGUGGAGGUUCGUCUAAUGACUA -...((((....))))......((((((.((((((.....))))))(((..((((..((((.((((((.(((((....))))).)))))).))))..))))..)))))))))........ ( -48.10) >DroMoj_CAF1 3366 106 - 1 -CAUGGACUGGUAUCCCGAGGAGGAUGAGGAGGACUUGAUGUUCUCGCCGGCCCUGUUGGCGCGACGGGCCAGCGAAAGCUGGAUAGUGGAGCCGCCCGUUGAGGUG------------- -((.((.(((((((((......))))...((((((.....))))))))))))).))..(((.(.((...(((((....)))))...)).).)))(((......))).------------- ( -44.40) >DroAna_CAF1 1886 109 - 1 CCAUGGAUUGGUAUCCGGAGGAGGAUGAGGAGGACCUCAUGUUCUCGCCAGCUCUGCUGGCCAGGCGGGCCAGUGAAAGCUGGAUAGUUGAACCACCAGUUGAGGUGGC----------- ...(((((....)))))(((((.(.(((((....)))))).))))).((((((..(((((((.....)))))))...)))))).........(((((......))))).----------- ( -47.40) >consensus _CAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACCUGAUGUUCUCGCCGGCGCUGUUGGCGCGACGAGCCAGCGAAAGCUGGAUAGUUGAGCCGCCCGUUGAGGUG_____________ .......(((((((((......))))...((((((.....))))))))))).......(((.((((...(((((....)))))...)))).)))(((......))).............. (-31.55 = -32.05 + 0.50)

| Location | 9,550,502 – 9,550,597 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -29.88 |
| Consensus MFE | -17.60 |
| Energy contribution | -18.02 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798733 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9550502 95 + 22407834 GCUUUCGCUGGCUCUUCUGGCCAACAAAGCCGGCGAGAACAUGAGGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUGCUCGCUGAU---UUUCUAC--- (((((.(.(((((.....))))).))))))(((((((...((((((....))))))........((((....))))...)))))))..---.......--- ( -33.30) >DroPse_CAF1 3141 95 + 1 GCUCUCGCUGGCACGACGCGCCAACAGAGCGGGCGAGAACAUUAGGUCCUCCUCAUCCUCCUCGGGAUACCAGUCCAUUGUGU---GC---UUUCGAUUUC (((((.(.((((.......)))).))))))(((((((.((.....)).)))...(((((....)))))....)))).......---..---.......... ( -26.30) >DroGri_CAF1 3698 95 + 1 ACUUUCACUAGCGCGACGCGCCAACAACGCCGGCGAGAACAUCAAGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUGUUGUUCU-U---CUU--GUUGU ..........((((...))))..((((((..((.(((((((.((.(..((....((((......))))...))..).)).))))))-)---)))--))))) ( -21.90) >DroEre_CAF1 1657 98 + 1 GCUUUCGCUGGCUCUCCUGGCCAGCAGCGCCGGCGAGAACAUGAGGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUGCUAGCCGCU---CUUCAACUUG ......(((((((.....)))))))(((((..(((((...((((((....)))))).)))....((((....))))..))..).))))---.......... ( -35.30) >DroYak_CAF1 310 101 + 1 GCUUUCGCUGGCUCGUCUGGCCAACAAGGCCGGCGAGAACAUGAGGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUUCUCGACUAUAGCAAUCAACUGG ...((((..((((((.((((((.....))))))))))...((((((....))))))...))..))))..(((((..(((((.......)).)))..))))) ( -33.10) >DroMoj_CAF1 3393 90 + 1 GCUUUCGCUGGCCCGUCGCGCCAACAGGGCCGGCGAGAACAUCAAGUCCUCCUCAUCCUCCUCGGGAUACCAGUCCAUGCUGACCC-C---UUU------- (.((((((((((((............)))))))))))).)..............(((((....)))))..((((....))))....-.---...------- ( -29.40) >consensus GCUUUCGCUGGCUCGUCGCGCCAACAAAGCCGGCGAGAACAUGAGGUCCUCCUCAUCCUCCUCCGGAUACCAGUCCAUGCUGGCCG_U___UUUC_ACU__ ..(((((((((((..............)))))))))))..........................((((....))))......................... (-17.60 = -18.02 + 0.42)


| Location | 9,550,502 – 9,550,597 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 77.96 |
| Mean single sequence MFE | -34.68 |
| Consensus MFE | -24.77 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.61 |
| SVM RNA-class probability | 0.798837 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9550502 95 - 22407834 ---GUAGAAA---AUCAGCGAGCAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACCUCAUGUUCUCGCCGGCUUUGUUGGCCAGAAGAGCCAGCGAAAGC ---.......---((((((.......).)))))..((((.((((.((((((....)))))).))))..)))).((((((((((....).)))))))))... ( -33.30) >DroPse_CAF1 3141 95 - 1 GAAAUCGAAA---GC---ACACAAUGGACUGGUAUCCCGAGGAGGAUGAGGAGGACCUAAUGUUCUCGCCCGCUCUGUUGGCGCGUCGUGCCAGCGAGAGC .....(....---).---.......((.....(((((......)))))..((((((.....))))))..))(((((((((((((...)))))))).))))) ( -30.90) >DroGri_CAF1 3698 95 - 1 ACAAC--AAG---A-AGAACAACAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACUUGAUGUUCUCGCCGGCGUUGUUGGCGCGUCGCGCUAGUGAAAGU .....--...---.-....(((((..(.(((((((((......))))...((((((.....))))))))))))..)))))((((...)))).......... ( -28.50) >DroEre_CAF1 1657 98 - 1 CAAGUUGAAG---AGCGGCUAGCAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACCUCAUGUUCUCGCCGGCGCUGCUGGCCAGGAGAGCCAGCGAAAGC ...(((....---)))((((..(.(((.(..(((.((((.((((.((((((....)))))).))))..))))...)))..)))).)..)))).((....)) ( -41.20) >DroYak_CAF1 310 101 - 1 CCAGUUGAUUGCUAUAGUCGAGAAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACCUCAUGUUCUCGCCGGCCUUGUUGGCCAGACGAGCCAGCGAAAGC ((((((.(((.((.......))))).))))))...((((.((((.((((((....)))))).))))..))))..(((((((((....).)))))))).... ( -38.90) >DroMoj_CAF1 3393 90 - 1 -------AAA---G-GGGUCAGCAUGGACUGGUAUCCCGAGGAGGAUGAGGAGGACUUGAUGUUCUCGCCGGCCCUGUUGGCGCGACGGGCCAGCGAAAGC -------...---.-(.(((((((.((.(((((((((......))))...((((((.....))))))))))))).))))))).).........((....)) ( -35.30) >consensus __AGU_GAAA___A_AGGCCAGCAUGGACUGGUAUCCGGAGGAGGAUGAGGAGGACCUCAUGUUCUCGCCGGCCCUGUUGGCCAGACGAGCCAGCGAAAGC .........................((.(((((((((......))))...((((((.....))))))))))))).(((((((.......)))))))..... (-24.77 = -24.75 + -0.02)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:22 2006