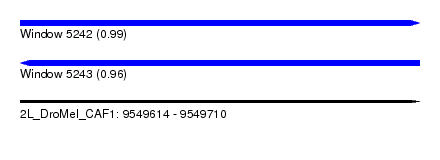
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,549,614 – 9,549,710 |
| Length | 96 |
| Max. P | 0.985541 |

| Location | 9,549,614 – 9,549,710 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 67.30 |
| Mean single sequence MFE | -22.04 |
| Consensus MFE | -14.28 |
| Energy contribution | -14.95 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.01 |
| SVM RNA-class probability | 0.985541 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9549614 96 + 22407834 CACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGAUGG--AUAAACAACAUUG----AAUUAUAUUCAAAAU-------AUGGUGUUA (((((((...((((((((.((((((....))))))((........)).))))).)))..--.....)))).(((----(((...))))))...-------...)))... ( -26.10) >DroVir_CAF1 2779 81 + 1 CAUAUUGACAAAAAGCACGAGCAGCAUCAGCUGUCGUCGCACCAGCCAAUCCUGUCCAGAAAUAAAUGAAGUAG----CAUU-----UA--G----------------- ......((((....(.((((.((((....)))))))))((....))......))))......((((((......----))))-----))--.----------------- ( -15.90) >DroGri_CAF1 2899 83 + 1 CACAUUGACAAACAGCACGAGCAGCAUCAGCUGUUGGCGCACAAACCAAUCCUGUCAA---AGAAAUUA---------UAAU-----UA--GU-------UGGACGAUA ...((((.......((.(.((((((....)))))).).)).....(((((.......(---(....)).---------....-----..--))-------))).)))). ( -18.04) >DroSec_CAF1 37 75 + 1 CACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGACGCACAAACCAGUCCUGGAUA---AUAAACAACAUUG----AAUU--------------------------- ...((((....(((((((.((((((....))))))(.....)......)))))))...---.....))))....----....--------------------------- ( -19.50) >DroSim_CAF1 37 95 + 1 CACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGAUA---AUAAACAACAUUG----AAUUAUAUUAAAAAU-------AUGGUGUUA ...(((.....(((((((.((((((....))))))((........)).)))))))...---...)))(((((((----.(((........)))-------.))))))). ( -25.20) >DroEre_CAF1 625 106 + 1 CACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGGUG---AUAAACAGCAUUCGUUCGAUUAGAUUCAAAUUGCAUUCCAUUGAAGCA ....((((((((((((((.((((((....))))))((........)).)))))(..((---(..........)))..)))))...)))))..(((.(((....)))))) ( -27.50) >consensus CACGUUGAAAAUCAGGACGAGCAGCAUCAGCUGCUGGCGCACAAACCAGUCCUGGAUA___AUAAACAACAUUG____AAUU_____CA__AU_______AUGG_G_UA ...........(((((((.((((((....))))))((........)).)))))))...................................................... (-14.28 = -14.95 + 0.67)

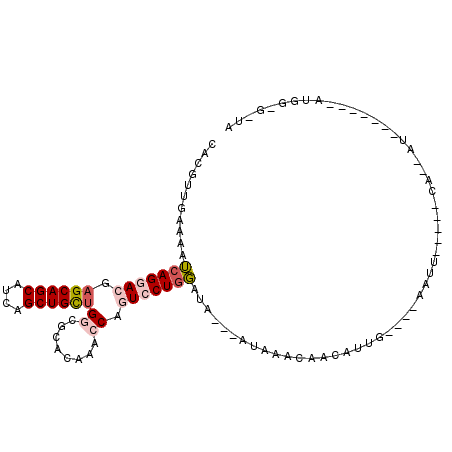
| Location | 9,549,614 – 9,549,710 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 67.30 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -14.86 |
| Energy contribution | -15.25 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.62 |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.956501 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9549614 96 - 22407834 UAACACCAU-------AUUUUGAAUAUAAUU----CAAUGUUGUUUAU--CCAUCCAGGACUGGUUUGUGCGCCAGCAGCUGAUGCUGCUCGUCCUGAUUUUCAACGUG ...(((...-------...((((((...)))----))).((((...((--(.....(((((((((......))))(((((....)))))..))))))))...))))))) ( -26.10) >DroVir_CAF1 2779 81 - 1 -----------------C--UA-----AAUG----CUACUUCAUUUAUUUCUGGACAGGAUUGGCUGGUGCGACGACAGCUGAUGCUGCUCGUGCUUUUUGUCAAUAUG -----------------.--((-----((((----......))))))........((..((((((.((..(((.(.((((....))))))))..))....)))))).)) ( -21.50) >DroGri_CAF1 2899 83 - 1 UAUCGUCCA-------AC--UA-----AUUA---------UAAUUUCU---UUGACAGGAUUGGUUUGUGCGCCAACAGCUGAUGCUGCUCGUGCUGUUUGUCAAUGUG .........-------..--..-----....---------........---((((((((.(((((......)))))(((((((......))).)))))))))))).... ( -16.50) >DroSec_CAF1 37 75 - 1 ---------------------------AAUU----CAAUGUUGUUUAU---UAUCCAGGACUGGUUUGUGCGUCAGCAGCUGAUGCUGCUCGUCCUGAUUUUCAACGUG ---------------------------....----..((((((.....---.(((.(((((.(((....(((((((...))))))).))).))))))))...)))))). ( -25.10) >DroSim_CAF1 37 95 - 1 UAACACCAU-------AUUUUUAAUAUAAUU----CAAUGUUGUUUAU---UAUCCAGGACUGGUUUGUGCGCCAGCAGCUGAUGCUGCUCGUCCUGAUUUUCAACGUG .........-------...............----..((((((.....---.(((.(((((((((......))))(((((....)))))..))))))))...)))))). ( -25.80) >DroEre_CAF1 625 106 - 1 UGCUUCAAUGGAAUGCAAUUUGAAUCUAAUCGAACGAAUGCUGUUUAU---CACCCAGGACUGGUUUGUGCGCCAGCAGCUGAUGCUGCUCGUCCUGAUUUUCAACGUG ((((((....))).)))..(((((.......(((((.....)))))..---....((((((((((......))))(((((....)))))..))))))...))))).... ( -28.80) >consensus UA_C_CCAU_______AC__UA_____AAUU____CAAUGUUGUUUAU___UAUCCAGGACUGGUUUGUGCGCCAGCAGCUGAUGCUGCUCGUCCUGAUUUUCAACGUG .......................................................((((((((((......))))(((((....)))))..))))))............ (-14.86 = -15.25 + 0.39)
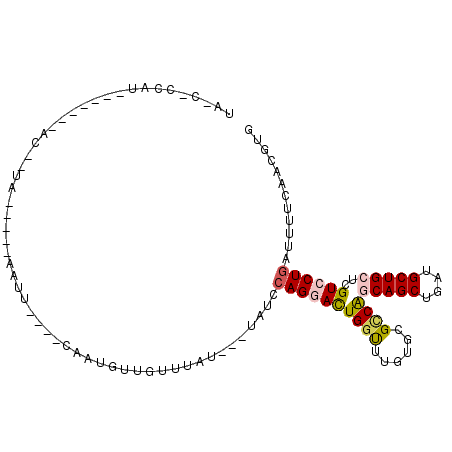
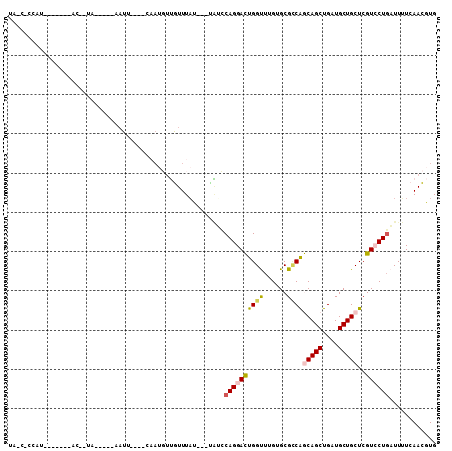
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:16 2006