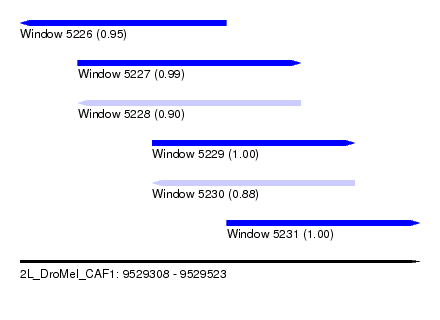
| Sequence ID | 2L_DroMel_CAF1 |
|---|---|
| Location | 9,529,308 – 9,529,523 |
| Length | 215 |
| Max. P | 0.998959 |

| Location | 9,529,308 – 9,529,419 |
|---|---|
| Length | 111 |
| Sequences | 4 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.67 |
| Mean single sequence MFE | -24.73 |
| Consensus MFE | -15.11 |
| Energy contribution | -14.37 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953227 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9529308 111 - 22407834 UCCCGCCAGAACUACCUGUUGGUUUUUCCCCAGGACUUACAAUCGGUUUCUCUUCGGGACGUCCUGUCGAGCCAUCGCUAUCACCUCCAGGUUUUCCUUUUGGAAUUUCUA .....((((((..(((((..(((...((..((((((...(....)((..(.....)..))))))))..))((....))....)))..))))).....))))))........ ( -28.10) >DroSec_CAF1 36820 101 - 1 UUCCGCCAGAACUACCUGCUGGUUUUCCUACAGGACUUCCAAUCGGGUUCUCUCCGGGACGUCCUUUCGAGCCAUCGCUAUCACCUCCAGGUUUUCCCUCU---------- ........(((..(((((..(((........(((((.(((...((((.....))))))).)))))....(((....)))...)))..))))).))).....---------- ( -26.30) >DroSim_CAF1 39930 92 - 1 UUCCGCCA---------GCUGGUUUUCCUCCAGGACUUCCAAUCGGGUUCUCUCCGGGACGUCCUUUCGAGCCAUCGCUAUCAGCUCCAGGUUUUCCCUCU---------- ..((...(---------((((((........(((((.(((...((((.....))))))).)))))....(((....))))))))))...))..........---------- ( -24.80) >DroYak_CAF1 44724 96 - 1 UUUCUCCAGAACUACCUGCAGGUUUUCCUCCAGGACUUCCAAUCGGGUUUUCUCCGAGUCGUCCUUUCGAUCCUUCACUAUCCCCCCCAGGAUUUC--------------- ..............((((.(((....))).))))...(((..(((((.....)))))((((......))))..................)))....--------------- ( -19.70) >consensus UUCCGCCAGAACUACCUGCUGGUUUUCCUCCAGGACUUCCAAUCGGGUUCUCUCCGGGACGUCCUUUCGAGCCAUCGCUAUCACCUCCAGGUUUUCCCUCU__________ ..(((.(((......))).))).........(((((......(((((.....)))))...)))))...(((((................)))))................. (-15.11 = -14.37 + -0.75)

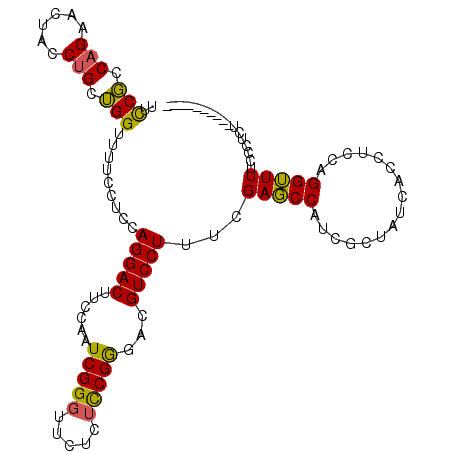
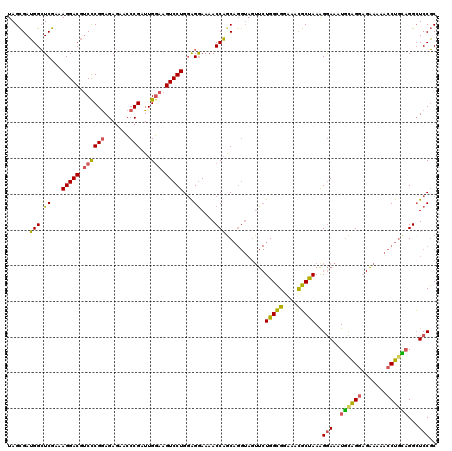
| Location | 9,529,339 – 9,529,459 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -39.65 |
| Consensus MFE | -25.85 |
| Energy contribution | -25.35 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.65 |
| SVM decision value | 2.09 |
| SVM RNA-class probability | 0.987695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9529339 120 + 22407834 UAGCGAUGGCUCGACAGGACGUCCCGAAGAGAAACCGAUUGUAAGUCCUGGGGAAAAACCAACAGGUAGUUCUGGCGGGAAUGCUAAGGGAAAUGUAGGAGAAAAACCUGCAGGCUCCGC (((((....((((.((((((.((((..((.((.((.....))...)))).))))...(((....))).)))))).))))..)))))..(((..((((((.......))))))...))).. ( -37.20) >DroSec_CAF1 36841 120 + 1 UAGCGAUGGCUCGAAAGGACGUCCCGGAGAGAACCCGAUUGGAAGUCCUGUAGGAAAACCAGCAGGUAGUUCUGGCGGAAACGCUAAAGGAAAUACAGGAGAAAAACCUGUAGGCUCCGC ....((((..((....)).)))).(((((.....((....((((.((((((.((....)).))))).).))))((((....))))...))...((((((.......))))))..))))). ( -43.00) >DroSim_CAF1 39951 108 + 1 UAGCGAUGGCUCGAAAGGACGUCCCGGAGAGAACCCGAUUGGAAGUCCUGGAGGAAAACCAGC---------UGGCGGAAACGCUAAAGAAAAUGCAGG---AAAACCUGUAGGCUCCGC ..(((....(((...(((((.((((((.......)))...))).))))).))).......(((---------(((((....))))........((((((---....)))))))))).))) ( -37.80) >DroYak_CAF1 44740 120 + 1 UAGUGAAGGAUCGAAAGGACGACUCGGAGAAAACCCGAUUGGAAGUCCUGGAGGAAAACCUGCAGGUAGUUCUGGAGAAAACUCCAAAGGAAGUGCGGUAGGAAAACCUACUGGGUCCUC ......((((((...(((((...((((.......))))........((((.(((....))).))))..)))))((((....))))..........(((((((....))))))))))))). ( -40.60) >consensus UAGCGAUGGCUCGAAAGGACGUCCCGGAGAGAACCCGAUUGGAAGUCCUGGAGGAAAACCAGCAGGUAGUUCUGGCGGAAACGCUAAAGGAAAUGCAGGAGAAAAACCUGCAGGCUCCGC ......(((.((...(((((.((((((.......)))...))).)))))...))....)))...........(((((....)))))..(((..((((((.......))))))...))).. (-25.85 = -25.35 + -0.50)

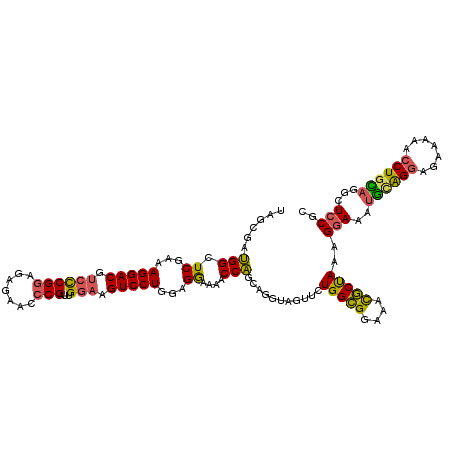
| Location | 9,529,339 – 9,529,459 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.83 |
| Mean single sequence MFE | -34.55 |
| Consensus MFE | -21.24 |
| Energy contribution | -20.99 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.898648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9529339 120 - 22407834 GCGGAGCCUGCAGGUUUUUCUCCUACAUUUCCCUUAGCAUUCCCGCCAGAACUACCUGUUGGUUUUUCCCCAGGACUUACAAUCGGUUUCUCUUCGGGACGUCCUGUCGAGCCAUCGCUA ((((((((.((((((..((((...............((......)).))))..)))))).)))))(((..((((((...(....)((..(.....)..))))))))..)))....))).. ( -31.99) >DroSec_CAF1 36841 120 - 1 GCGGAGCCUACAGGUUUUUCUCCUGUAUUUCCUUUAGCGUUUCCGCCAGAACUACCUGCUGGUUUUCCUACAGGACUUCCAAUCGGGUUCUCUCCGGGACGUCCUUUCGAGCCAUCGCUA ..((((..((((((.......)))))).))))..(((((.....((.((......))))((((((......(((((.(((...((((.....))))))).)))))...)))))).))))) ( -35.10) >DroSim_CAF1 39951 108 - 1 GCGGAGCCUACAGGUUUU---CCUGCAUUUUCUUUAGCGUUUCCGCCA---------GCUGGUUUUCCUCCAGGACUUCCAAUCGGGUUCUCUCCGGGACGUCCUUUCGAGCCAUCGCUA ((((((((((((((....---)))).........))).).)))))).(---------((((((((......(((((.(((...((((.....))))))).)))))...))))))..))). ( -33.10) >DroYak_CAF1 44740 120 - 1 GAGGACCCAGUAGGUUUUCCUACCGCACUUCCUUUGGAGUUUUCUCCAGAACUACCUGCAGGUUUUCCUCCAGGACUUCCAAUCGGGUUUUCUCCGAGUCGUCCUUUCGAUCCUUCACUA ((((((((.(((((....))))).........(((((((....)))))))....((((.(((....))).))))..........))))))))...(..(((......)))..)....... ( -38.00) >consensus GCGGAGCCUACAGGUUUUUCUCCUGCAUUUCCUUUAGCGUUUCCGCCAGAACUACCUGCUGGUUUUCCUCCAGGACUUCCAAUCGGGUUCUCUCCGGGACGUCCUUUCGAGCCAUCGCUA ((((((((((.(((................))).))).))).))))...........(.((((((......(((((......(((((.....)))))...)))))...)))))).).... (-21.24 = -20.99 + -0.25)

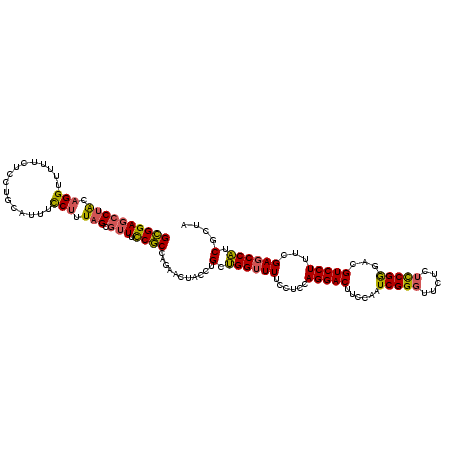
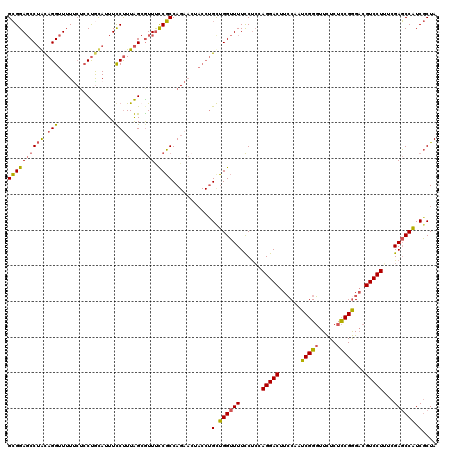
| Location | 9,529,379 – 9,529,488 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -25.85 |
| Energy contribution | -24.20 |
| Covariance contribution | -1.65 |
| Combinations/Pair | 1.36 |
| Mean z-score | -3.61 |
| Structure conservation index | 0.73 |
| SVM decision value | 3.28 |
| SVM RNA-class probability | 0.998917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9529379 109 + 22407834 GUAAGUCCUGGGGAAAAACCAACAGGUAGUUCUGGCGGGAAUGCUAAGGGAAAUGUAGGAGAAAAACCUGCAGGCUCCGCUGGAGGAACCACCAACACAAAUAUCAACA ....((..(((((.....))....(((..(((..(((((...(((........((((((.......))))))))))))))..)))..))).)))..))........... ( -30.00) >DroSec_CAF1 36881 109 + 1 GGAAGUCCUGUAGGAAAACCAGCAGGUAGUUCUGGCGGAAACGCUAAAGGAAAUACAGGAGAAAAACCUGUAGGCUCCGCUGGAGGAACCACCAACAAAAAUAUCAACA ((((.((((((.((....)).))))).).))))((((....))))...((...((((((.......))))))((.(((......))).)).))................ ( -36.00) >DroYak_CAF1 44780 109 + 1 GGAAGUCCUGGAGGAAAACCUGCAGGUAGUUCUGGAGAAAACUCCAAAGGAAGUGCGGUAGGAAAACCUACUGGGUCCUCUGAAGGAUCCAAGAACGUAAAUAGCAACG ((((.(((((.(((....))).)))).).))))((((....))))........(((((((((....))))))(((((((....))))))).............)))... ( -39.60) >consensus GGAAGUCCUGGAGGAAAACCAGCAGGUAGUUCUGGCGGAAACGCUAAAGGAAAUGCAGGAGAAAAACCUGCAGGCUCCGCUGGAGGAACCACCAACAAAAAUAUCAACA .....((((..((((..(((....)))..))))((((....))))..))))...(((((.......))))).((.(((......))).))................... (-25.85 = -24.20 + -1.65)

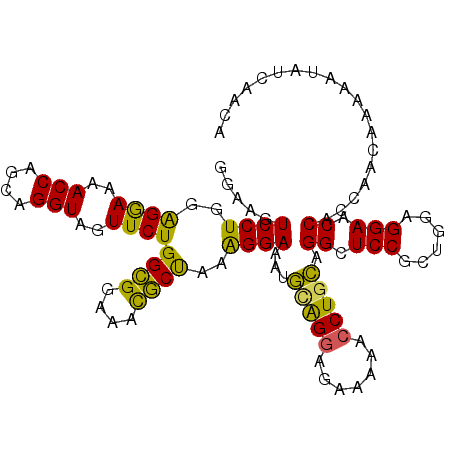
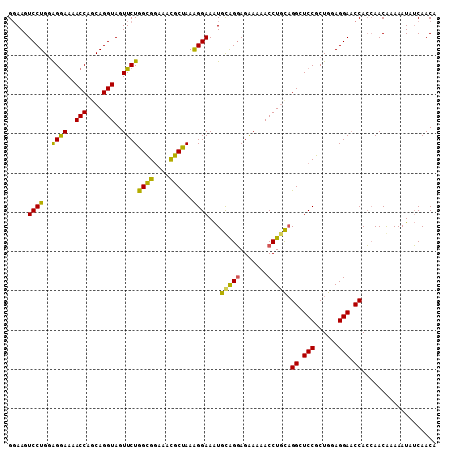
| Location | 9,529,379 – 9,529,488 |
|---|---|
| Length | 109 |
| Sequences | 3 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.12 |
| Mean single sequence MFE | -30.23 |
| Consensus MFE | -18.23 |
| Energy contribution | -19.12 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.882163 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9529379 109 - 22407834 UGUUGAUAUUUGUGUUGGUGGUUCCUCCAGCGGAGCCUGCAGGUUUUUCUCCUACAUUUCCCUUAGCAUUCCCGCCAGAACUACCUGUUGGUUUUUCCCCAGGACUUAC .(((.........(((((.(....).)))))((((((.((((((..((((...............((......)).))))..)))))).)))...)))....))).... ( -24.59) >DroSec_CAF1 36881 109 - 1 UGUUGAUAUUUUUGUUGGUGGUUCCUCCAGCGGAGCCUACAGGUUUUUCUCCUGUAUUUCCUUUAGCGUUUCCGCCAGAACUACCUGCUGGUUUUCCUACAGGACUUCC .(((((.......(((((.(....).)))))((((..((((((.......)))))).)))).)))))((((......))))..((((..((....))..))))...... ( -30.00) >DroYak_CAF1 44780 109 - 1 CGUUGCUAUUUACGUUCUUGGAUCCUUCAGAGGACCCAGUAGGUUUUCCUACCGCACUUCCUUUGGAGUUUUCUCCAGAACUACCUGCAGGUUUUCCUCCAGGACUUCC .............(((((.(((..((((((((((..(.(((((....))))).)....))))))))))..)))...)))))..((((.(((....))).))))...... ( -36.10) >consensus UGUUGAUAUUUAUGUUGGUGGUUCCUCCAGCGGAGCCUGCAGGUUUUUCUCCUGCAUUUCCUUUAGCGUUUCCGCCAGAACUACCUGCUGGUUUUCCUCCAGGACUUCC ...................((.((((..((..(((((.((((((..((((...............(((....))).))))..)))))).)))))..))..))))))... (-18.23 = -19.12 + 0.89)

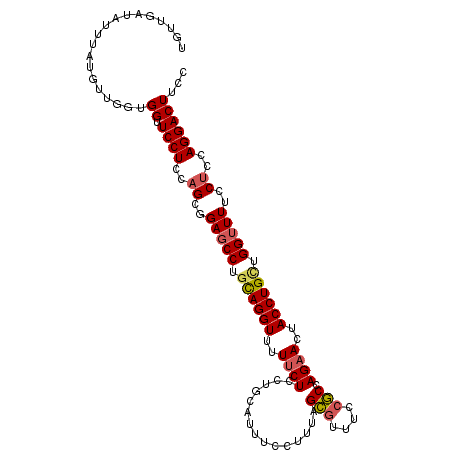
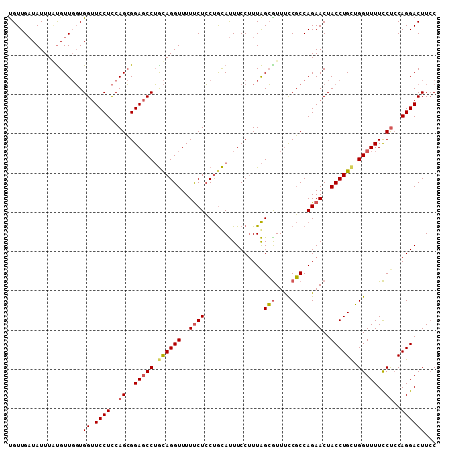
| Location | 9,529,419 – 9,529,523 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.12 |
| Mean single sequence MFE | -20.75 |
| Consensus MFE | -14.16 |
| Energy contribution | -14.10 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.68 |
| SVM decision value | 3.30 |
| SVM RNA-class probability | 0.998959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2L_DroMel_CAF1 9529419 104 + 22407834 AUGCUAAGGGAAAUGUAGGAGAAAAACCUGCAGGCUCCGCUGGAGGAACCACCAACACAAAUAUCAACACUAUUAAAAUACAACACAAACGCUAACUUUAGAAA----- ...((((((....((((((.......))))))((.(((......))).)).............................................))))))...----- ( -18.90) >DroSec_CAF1 36921 109 + 1 ACGCUAAAGGAAAUACAGGAGAAAAACCUGUAGGCUCCGCUGGAGGAACCACCAACAAAAAUAUCAACAUUAUUAAAAUACAAGACAAACGCUAACUUUAAAAGUAAAC ..((....((...((((((.......))))))((.(((......))).)).))......((((.......))))................))................. ( -16.40) >DroSim_CAF1 40022 100 + 1 ACGCUAAAGAAAAUGCAGG---AAAACCUGUAGGCUCCGCUGGAGGAACCACCAACAA-AAUAUCAACAAUAUUAAAAUACAACACAAACGCUAACUUUAGAAG----- ...((((((....((((((---....))))))((.(((......))).))........-....................................))))))...----- ( -19.30) >DroYak_CAF1 44820 94 + 1 ACUCCAAAGGAAGUGCGGUAGGAAAACCUACUGGGUCCUCUGAAGGAUCCAAGAACGUAAAUAGCAACGAUAUUAUUAUA---------GGCUGACUUUGAAA------ ....(((((....(((((((((....)))))((((((((....))))))))....))))..((((....(((....))).---------.)))).)))))...------ ( -28.40) >consensus ACGCUAAAGGAAAUGCAGGAGAAAAACCUGCAGGCUCCGCUGGAGGAACCACCAACAAAAAUAUCAACAAUAUUAAAAUACAACACAAACGCUAACUUUAAAAG_____ ...((((((....((((((.......))))))((.(((......))).)).............................................))))))........ (-14.16 = -14.10 + -0.06)

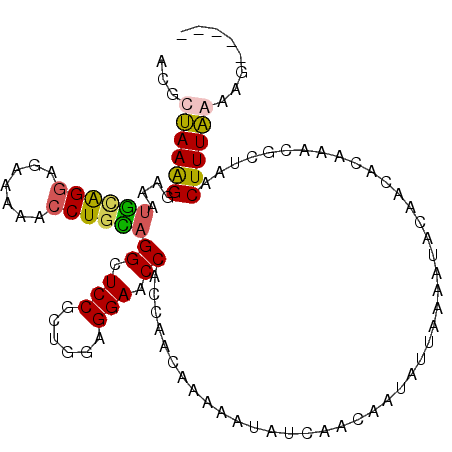
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:49:04 2006